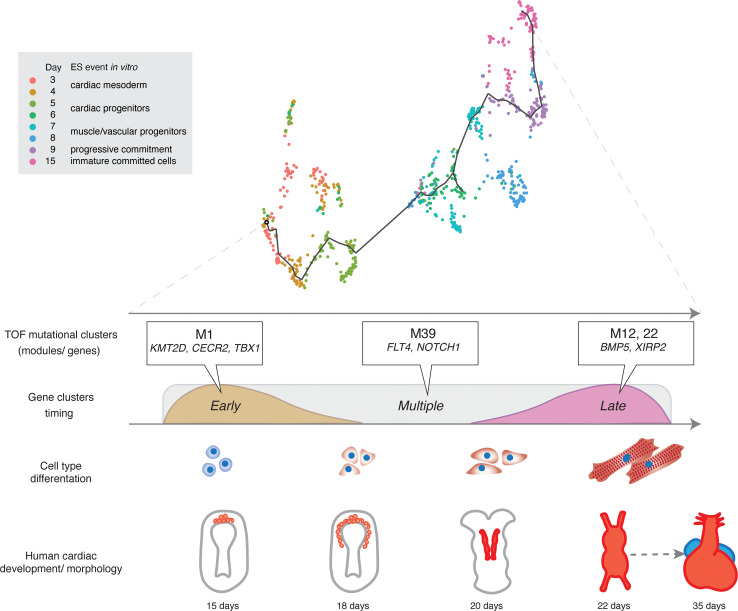
Figure 7. The genetic heterogeneity of TOF is reflected in a diversity of genetic modules, signaling pathways, and developmental timing of effect.
Intersection of human cardiac myocardial cell differentiation single-cell RNA-sequencing data set (top) with the mutational landscape of large cohorts of patients with TOF enabled identification of specific genes and gene modules significantly associated with distinct developmental time points (middle). Some genes such as FLT4 or NOTCH1 identified as having damaging variants in multiple sequencing studies are expressed at multiple developmental time points, whereas others are significantly constrained to either early (e.g., KMT2D) or late (e.g., BMP5) time points. Combining sequencing data with single-cell data sets provides new insight into the developmental mechanisms underlying the pathogenesis of TOF.