Figure 5.
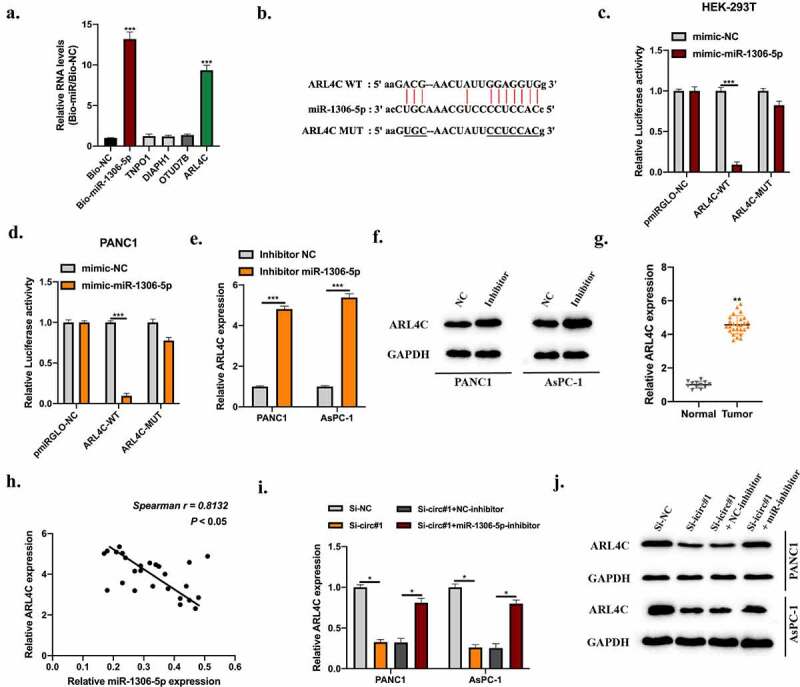
MiR-1306-5p directly targets ARL4C. Bioinformatics analysis were conducted using miRanDa (http://www.mirbase.org), miRmap (https://mirmap.ezlab.org), and TargetScan (http://www.targetscan.org/mamm_31) with CLIP data: strict stringency (≥5), Degradome data: low stringency (≥1). Putative targets were selected. A: RNA pull-down using Bio-NC probes and Bio-miR-1306-5p probes to detect putative mRNAs’ level. b: The predicted binding sites between miR-1306-5p and ARL4C were shown. c-d: Relative luciferase activities in HEK-293 T and PANC1 cells transfected with miR-1306-5p mimic and reporter vectors harboring WT or Mut ARL4C sequences were measured. e-f: ARL4C levels in PANC1 and AsPC-1 cells stably transfected with miR-1306-5p inhibitors, and thier normal control was measured by qRT-PCR and Western blot assays. g: ARL4C levels in adjacent normal tissues (n = 12) and PDAC tissues (n = 26) were measured by qRT-PCR. H: The expression correlation between miR-1306-5p and ARL4C in PDAC tissues was calculated by spearman analysis. i-j: ARL4C expression in Si-NC, Si-circ#1, Si-circ#1 + NC-inhibitor, Si-circ#1 + miR-1306-5p-inhibitor transfected PANC1, and AsPC-1 cells were measured by RT-qPCR and Western blot. All experiments were performed three times, and data were presented as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001.
