Figure 1.
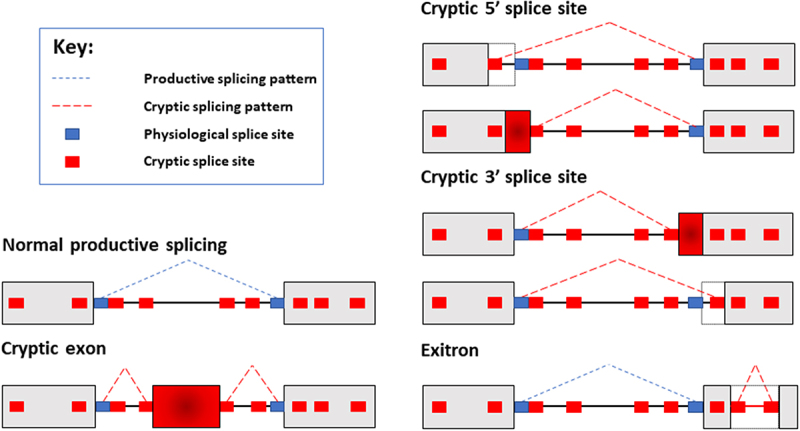
Schematic diagram of cryptic splicing patterns. Most genes are split between exons (shown as gray boxes here) and introns (shown as connecting lines between the boxes). Normal patterns of splice site selection will involve the spliceosome recognizing bona fide splice sites, and joining exons together to create mRNAs. In this example, normal productive splicing is indicated with dashed blue lines. Cryptic splice sites (smaller red boxes) resemble physiological splice sites (smaller blue boxes), and are found within both introns and exons. While normally these cryptic splice sites are ignored by the spliceosome, potentially they could act as decoy sites splice sites for spliceosome selection. Use of cryptic splice sites would produce different mRNAs from genes. Here the normal splicing patterns is shown as a broken blue line joining the physiological splice sites. Examples of cryptic splicing are indicated with dashed red lines. These cryptic splicing events are inclusion of a cryptic exon embedded deep within an intron; selection of cryptic 5ʹ and 3ʹ splice sites; and aberrant recognition of cryptic splice sites within an exon, leading to the interior of this exon being aberrantly recognized as an intron (in a cryptic splicing event known as an exitron).
