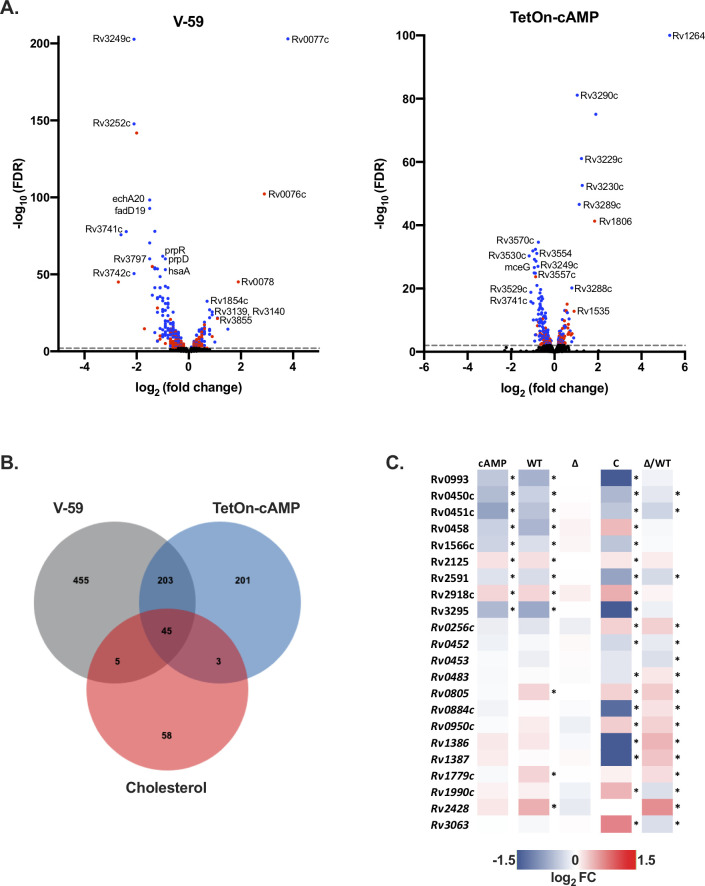
Fig 8. V-59 treatment and induction of TetOn-cAMP are associated with transcriptional changes in select CRPMt regulon genes.
(A) Volcano plots displaying differentially expressed genes following V-59 treatment of WT Mtb relative to DMSO control (left), or following Atc treatment of TetOn-cAMP Mtb relative to EtOH control (right), based on RNA-seq. Each dot represents a single gene, genes in blue are significant (FDR < 0.05) in both data sets and genes in red are unique to their respective data set. Dashed line indicates FDR cutoff < 0.01. (B) Venn diagram showing the number of significantly differentially expressed genes shared by the V-59 and TetOn-cAMP conditions, and how many of these belong to the KstR cholesterol-related regulon. (C) RNA-seq analysis quantifying differentially expressed genes from Mtb grown in cholesterol media, following V-59 treatment, or induction of TetOn-cAMP with Atc. Genes depicted are predicted members of the CRPMT regulon. Only genes with significant differential expression in both the V-59 and TetOn-cAMP conditions, or genes with intrinsic changes in the ∆Rv1625 strain (in italics), are shown. “cAMP” = Tet-On cAMP Atc vs. EtOH, “WT” = WT V-59 vs. DMSO, “∆” = ΔRv1625 V-59 vs. DMSO, “C” = CompFull V-59 vs. DMSO, “Δ/WT” = ∆Rv1625 DMSO vs. WT DMSO.