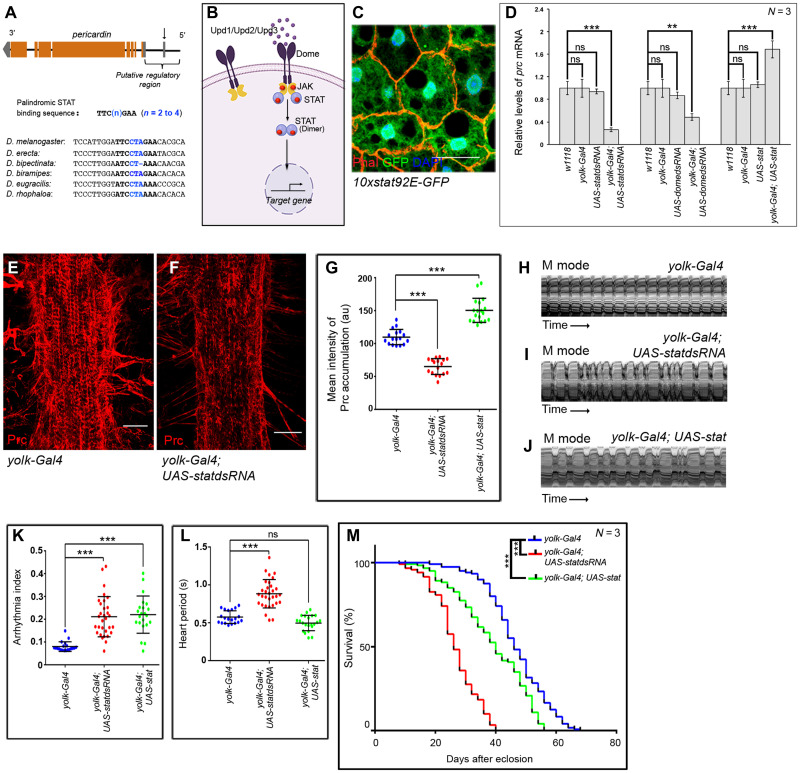
Fig. 2. JAK-STAT signaling regulates prc expression in the fat body cells.
(A) Schematic representation of prc genomic region (not up to scale) of Drosophila melanogaster, marking the potential STAT-binding site (arrow). Sequence alignment of the regulatory regions of the annotated genes coding for Collagen IV in different species of Drosophila highlighting the conserved potential STAT-binding site. (B) Schematic representation of the JAK-STAT signaling pathway in Drosophila. (C) Expression for 10xStat92EGFP (green) in adult fat cells. Phalloidin (red) marks the cell boundaries and DAPI (4′,6-diamidino-2-phenylindole) (blue) marks the nuclei. Scale bar, 20 μm. (D) Changes in prc expression upon modulating JAK-STAT signaling in fat cells. The transcript levels are normalized to that of the constitutive ribosomal gene rp49. (E and F) Down-regulating stat92E in fat cells reduces the level Prc accumulation around the second heart chamber (F) as compared to control (E). Scale bars, 25 μm. (G) Quantification of the mean fluorescence intensity for Prc accumulation around the second heart chamber upon modulating stat92E in fat cells. The dots represent the samples analyzed for each genotype. au, arbitrary units. (H to J) Representative M-mode records for heartbeats of adult flies showing the movement of the heart tube walls (y axis) over time (x axis). (K and L) Changes in Arrhythmia Index (K) and in Heart Period (L) upon modulating JAK-STAT signaling in adult fat cells. The dots represent the samples analyzed for each genotype. (M) Survival rates of yolk-Gal4; UAS-statdsRNA and yolk-Gal4; UAS-stat flies as compared to that of the control yolk-Gal4 flies. Genotypes are as mentioned. Data are represented as means ± SD. Statistical significance with P values of P < 0.01, and P < 0.001 are mentioned as **, and ***, respectively.