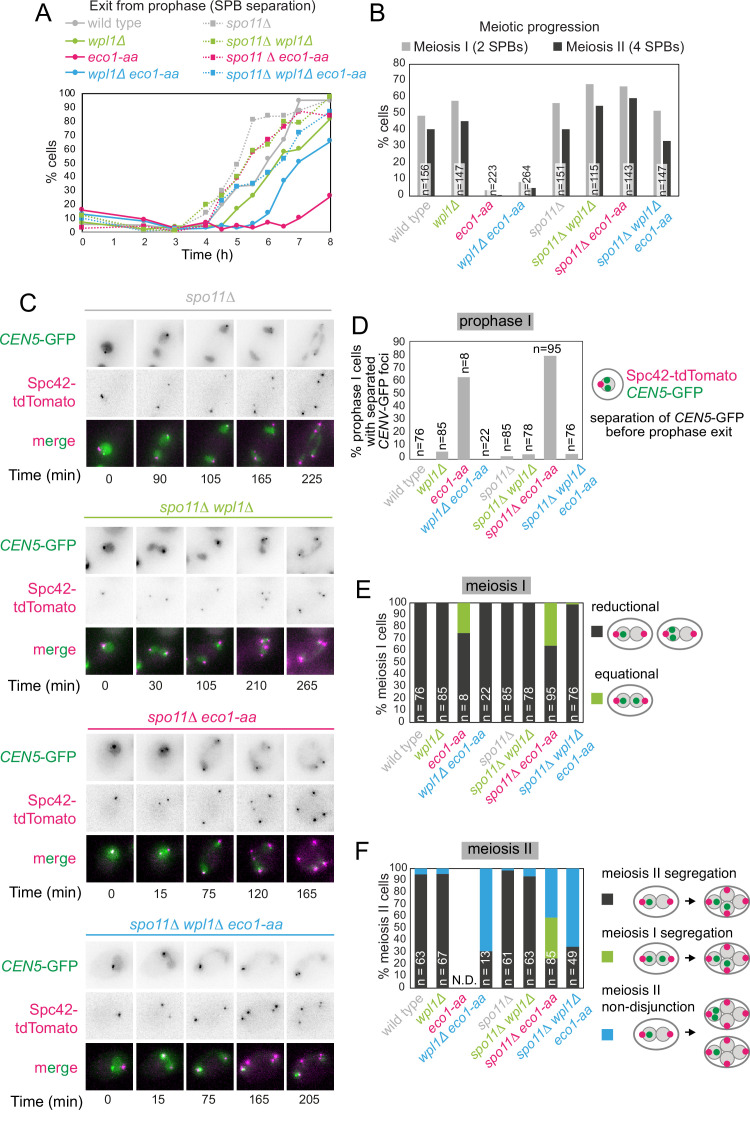
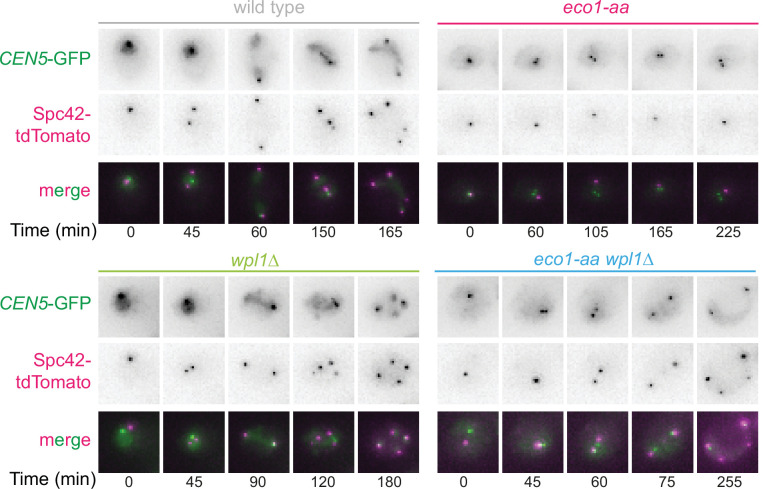
Figure 8. Critical Wpl1-dependent and independent roles of Eco1 in chromosome segregation during both meiosis I and II.
(A) Meiotic double-strand break formation arrests eco1-aa cells in meiotic prophase. Wild-type (AM24167), wpl1Δ (AM24168), eco1-aa (AM24184), eco1-aa wpl1Δ (AM24169), spo11Δ (AM27670) spo11Δ wpl1Δ (AM27673), spo11Δ eco1-aa (AM27672), and spo11Δ eco1-aa wpl1Δ (AM27671) anchor-away strains carrying heterozygous SPC42-tdTOMATO and CENV-GFP were induced to sporulate. The percentage of cells with more than one Spc42-tdTomato focus was determined from cell populations fixed at the indicated timepoints after resuspension in SPO medium containing rapamycin. At least 100 cells were scored per timepoint. (B–F) Live-cell imaging of strains as in (A) sporulated in the presence of rapamycin. Every cell that had one Spc42-tdTomato focus at the start of the movie was scored for the duration of the movie. A small number of mitotic/dead cells (<1% for each strain) were excluded from the analysis. (B) Deletion of SPO11 restores meiotic progression to eco1-aa cells. The percentage of cells that displayed two (meiosis I) or four (meiosis II) Spc42-tdTomato foci for each strain is shown. (C) Representative images of spo11Δ background cells undergoing the meiotic divisions. (D) Percentage of cells with one Spc42-tdTomato focus (prophase I) where two GFP foci were visible. Only cells that progressed to anaphase I were scored in this analysis. (E) Eco1 counteracts Wpl1 to allow the establishment of sister kinetochore mono-orientation. Segregation of CEN5-GFP foci to the same (reductional; dark gray) or opposite (equational; green) poles was scored in meiosis I (as two Spc42-tdTomato foci separate; binucleate cells). (F) Eco1 is required for pericentromeric cohesion, even in the absence of Wpl1. Segregation of CEN5-GFP foci to opposite (dark gray) or the same pole(s) (blue) was scored in meiosis II (as four Spc42-tdTomato foci separate; tetranucleate cells). Cells that had already segregated their sister CEN5-GFPs in meiosis I (GFP foci in two nuclei at the binucleate stage) were scored as a separate category (green).