Fig. 1. tRNAGly overexpression rescues inhibition of protein synthesis and peripheral neuropathy phenotypes in Drosophila CMT2D models.
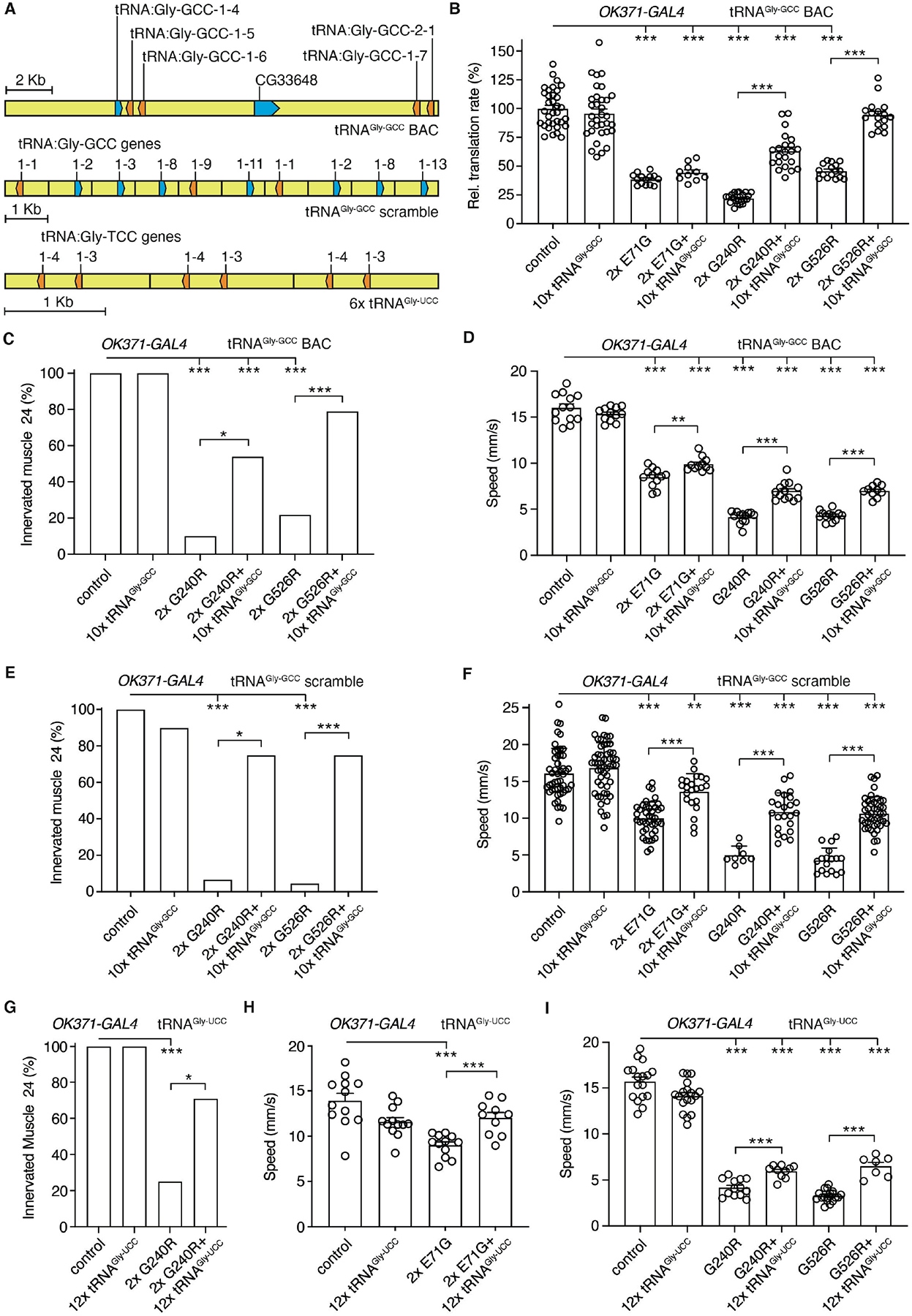
(A) Schematic of the transgenes used for tRNAGly-GCC or tRNAGly-UCC overexpression. (B) Relative translation rate as determined by FUNCAT in motor neurons (OK371-GAL4) of larvae expressing E71G, G240R, or G526R GlyRS (2x: two transgene copies), in the presence or absence of the tRNAGly-GCC BAC transgene (10xtRNAGly-GCC). n=10–34 animals per genotype; ***p<0.001 by Kruskal-Wallis test. (C,E,G) Percentage of larvae with innervated muscle 24. GlyRS transgenes were expressed in motor neurons (OK371-GAL4), in the presence or absence of 10xtRNAGly-GCC BAC (C), 10xtRNAGly-GCC scramble (E), or 12xtRNAGly-UCC (G). n=19–26 (C), 8–22 (E), 12–27 (G) animals per genotype; *p<0.05; ***p<0.005 by Fisher’s exact test with Bonferroni correction. (D,F,H,I) Negative geotaxis climbing speed of 7-day-old female flies expressing GlyRS transgenes in motor neurons (OK371-GAL4), in the presence or absence of 10xtRNAGly-GCC BAC (D), 10xtRNAGly-GCC scramble (F), or 12xtRNAGly-UCC (H,I). n=11–13 (D), 8–52 (F), 7–19 (H,I) groups of 10 flies per genotype; **p<0.01; ***p<0.005 by two-way ANOVA (D) or Brown-Forsythe and Welch ANOVA (F,H,I). Controls in (B-I) are driver-only. Graphs represent mean ± SEM.
