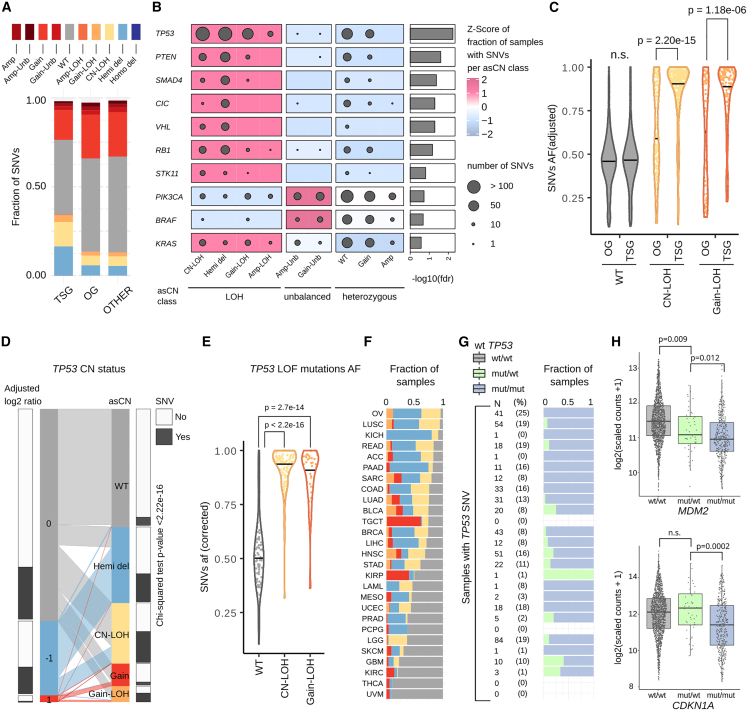
Figure 2.
Association between loss of heterozygosity, SNVs incidence, and their allelic fraction
(A) Fraction of SNVs (N = 601,177) in each asCN state, stratified by class of gene (TSG, OG, other genes). Color code is shared for (A, C, D, E, and F).
(B) Top-ten ranked genes with uneven distributions of SNVs across asCN classes. Dot size is proportional to the number of samples with SNV. Oncogenes and TSG with FDR < 0.001 (chi-square test) and with at least 20 SNV events are shown.
(C) Distribution of allelic fraction of SNV (N = 6,657, Mann-Whitney tests) across asCN classes comparing tumor suppressor genes and oncogenes.
(D) Parallel sets plot showing the mapping of TP53 log2 discretized copy-number values to asCN status. Barplots on the sides of CN bars show the fraction of samples harboring TP53 deleterious SNVs in each class. Chi-square test indicates the non-independence between asCN status and presence of SNVs.
(E) Distribution of allelic fractions of TP53 LOF mutations (N = 384, Mann-Whitney tests).
(F) Barplots showing the TP53 asCN status in each TCGA study.
(G) Fraction of samples with TP53 loss-of-function SNVs that have retained or lost the wild-type copy. Each row represents a TCGA study as indicated in (F).
(H) Expression levels of CDKN1A and MDM2 in samples stratified based on presence of wild-type copies of TP53.