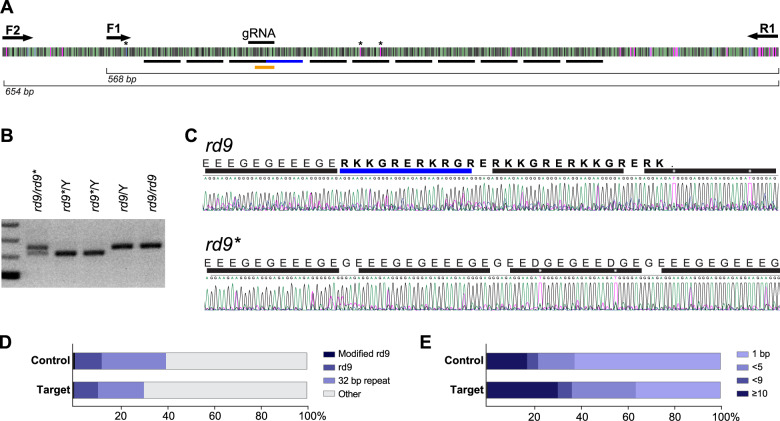
Fig. 6. RPGR-ORF15 sequencing following CRISPR modification.
A Schematic of the repetitive ORF15 sequence and primers used for PCR amplicon sequencing. Underlined black bars indicate locations of the 32-bp repeats and the underlined blue bar indicates the insertion in the rd9 mutant. Gene modifications were identified by mismatch within the region underlined in orange. Asterisks indicate unique base pairs used for alignments. B PCR strategy used to identify and sequence germline modified RPGR-ORF15 (rd9*). Amplification of the ORF15 locus in rd9* mice demonstrated a small deletion, evidenced by a smaller molecular weight band. C Sanger sequencing of the rd9* amplicon shown in (B). Protein coding sequence shown to highlight the restored reading frame in rd9* mice. Bars and asterisks are analogous to those depicted in panel (A). Correction in rd9* was identified as a precise 32-bp deletion. D Analysis of reads from PacBio deep sequencing of AAV ORF15-targeted retina and non-targeted control retina. Most sequences were degenerate with only 20–40% containing at least one precise 32-bp repeat (32 bp repeat). Only 11–12% of reads could be unambiguously mapped to the rd9 insertion region but very few (64 control; 33 ORF15 target) were identified as modified from native rd9 sequence (Modified rd9). The identification of modifications in both ORF15 and control-treated retina suggests these are sequencing errors. E Analysis of INDEL lengths identified in panel (D).