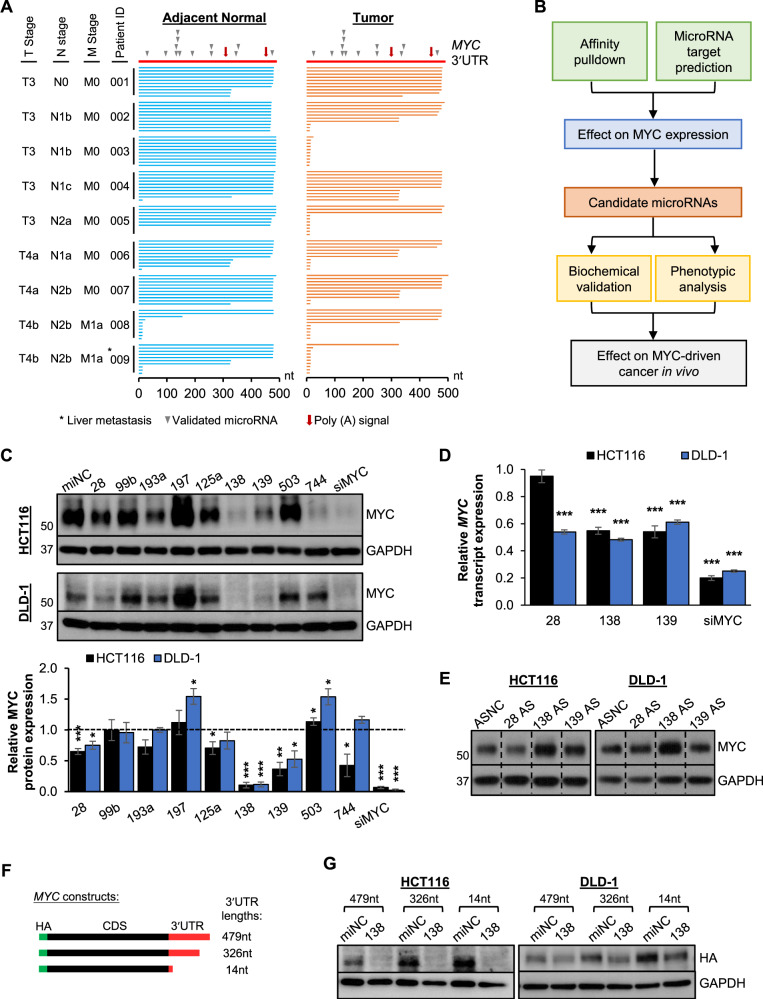
Fig. 1. Identification of potential MYC CDS-targeting miRNAs.
A Schematic representation of MYC 3′UTR isoforms detected by 3′ RACE in nine pairs of CRC tumor and adjacent normal patient samples (N = 9). B Flowchart outlining the workflow to identify and functionally characterize miRNAs that target the MYC CDS. MiRNAs were identified via affinity pulldown by MS2-tagged RNA Affinity Purification (MS2-TRAP) and miRNA target prediction by RNA22. C Western blot (top panel) and densitometry quantification (bottom panel) of MYC protein expression upon the overexpression of candidate miRNAs in HCT116 and DLD-1 cell lines. D RT-qPCR analysis of MYC transcript expression upon the overexpression of miR-28-5p, miR-138 and miR-139-3p in HCT116 and DLD-1 cell lines. E Western blot analysis of MYC protein expression upon miRNA inhibition by antisense miRNA inhibitors (AS) in HCT116 (left panel) and DLD-1 (right panel). F Schematic illustrating the HA-tagged MYC constructs with different 3′UTR lengths. G Western blot analysis of HA-tagged MYC protein expression upon miR-138 overexpression (138) in HCT116 (left panel) and DLD-1 (right panel). Mean ± SEM; N ≥ 3. *P < 0.05; **P < 0.01; ***P < 0.001.