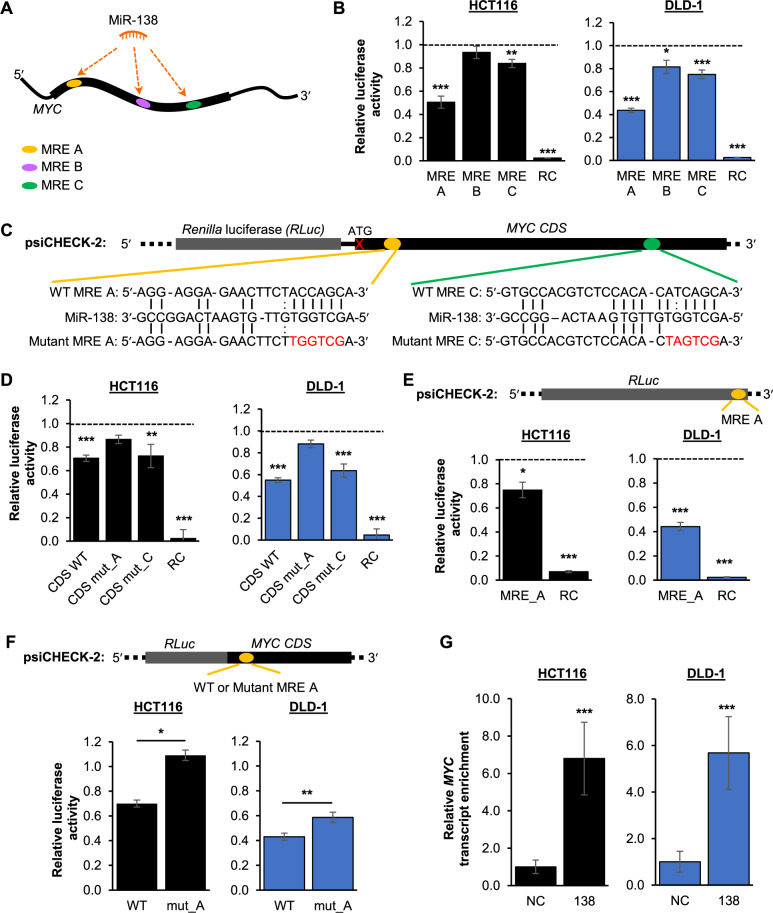
Fig. 2. Validation of miR-138 as a MYC CDS-targeting miRNA.
A Schematic representation of predicted miR-138 MREs on the MYC transcript. The thick line indicates the MYC CDS, while the thin lines indicate the UTRs. B Effect of miR-138 overexpression on the luciferase activity of the respective MRE-reporter constructs in HCT116 (left panel) and DLD-1 (right panel) cell lines. RC denotes the reverse complement of miR-138 which was used as positive control. C Schematic representation of the ATG-less MYC CDS cloned into the psiCHECK-2 vector. Location and sequences of the wild-type (WT) and mutant MREs A (yellow oval) and C (green oval) are depicted. Red font indicates the mutated nucleotides. The cross indicates the absence of the start codon ATG. D Effect of miR-138 overexpression on the luciferase activity of the MYC CDS reporter constructs with mutated MREs (CDS mut) compared to the wild-type CDS reporter (CDS WT) in HCT116 (left panel) and DLD-1 (right panel) cell lines. E Effect of miR-138 overexpression on the luciferase activity of the Renilla luciferase (RLuc) gene fused with MRE A in HCT116 (left panel) and DLD-1 (right panel) cell lines. Top panel illustrates the modification of RLuc gene construct. F Effect of miR-138 overexpression on the luciferase activity of the RLuc and MYC CDS fusion reporter construct with mutated MRE A (mut_A) compared to the wild-type MYC CDS (WT) in the HCT116 (left panel) and DLD-1 (right panel) cell lines. The top panel illustrates the fusion gene construct. G RT-qPCR analysis of MYC transcript enrichment upon biotinylated miR-138 pulldown in HCT116 (left panel) and DLD-1 (right panel) cell lines. (B, D, E and F) Mean ± SEM; N ≥ 3. G Mean ± STD; N ≥ 3. *P < 0.05; **P < 0.01; ***P < 0.001.