Fig 2. MD simulations of Cpx Ct21 peptides with lipid bilayers.
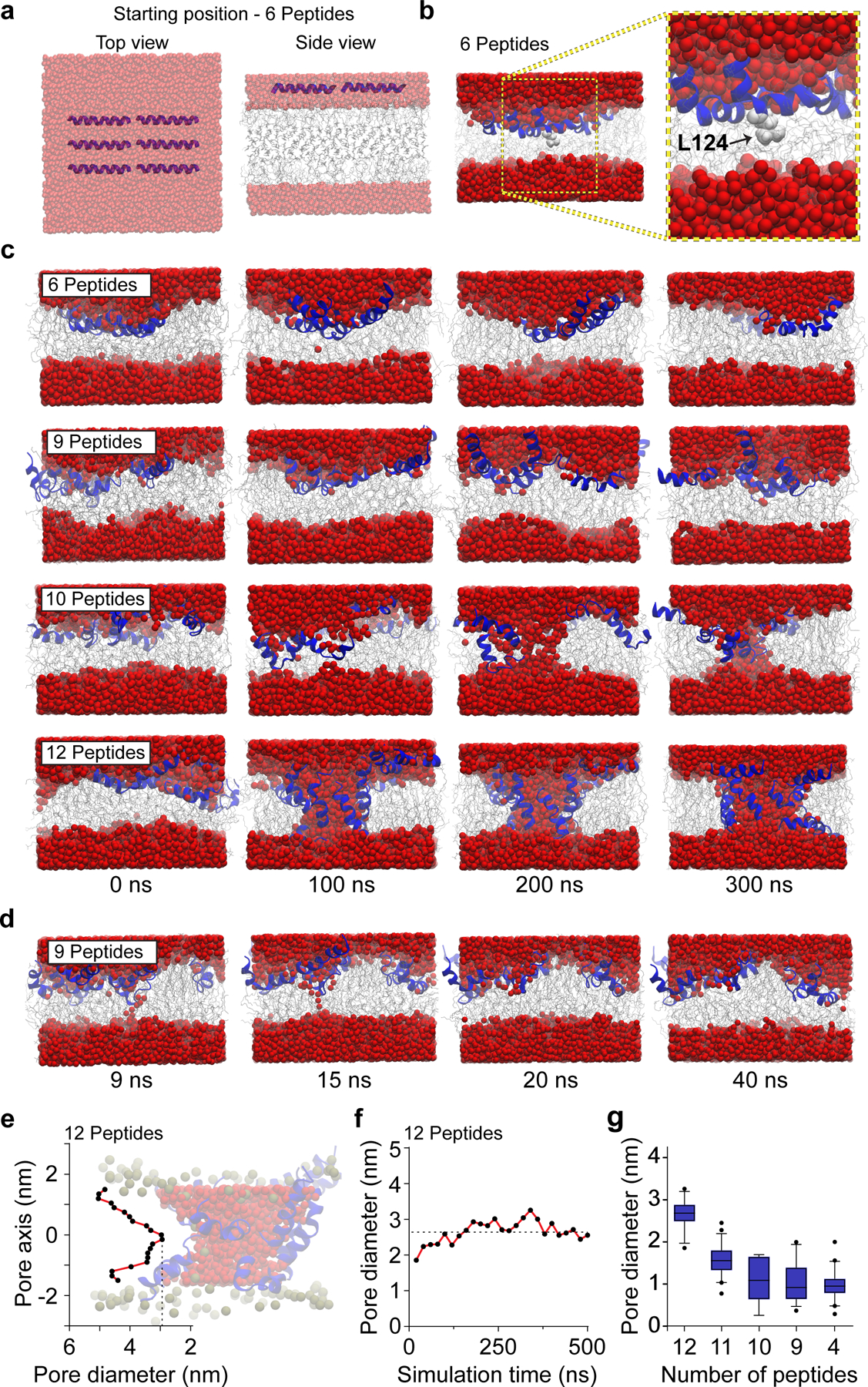
(a) HMMM model showing top and side views of six Ct21 peptides at the starting position of the MD simulation. Throughout the figure, water is shown as red spheres, lipids are grey, and Ct21 peptides are rendered in blue. Note: images in panel A are semi-transparent to aid in visualization of the Ct21 peptides within the layer of water molecules. (b) Six Ct21 peptide MD simulation snapshot and zoomed inset after 15 ns showing membrane insertion begins with L124 (light grey). (c) MD simulations snapshots between 0 and 300 ns with six, nine, ten or twelve Ct21 peptides. (d) MD simulation snapshots of nine Ct21 peptides in the bilayer, taken between 9 and 40 ns. Narrow transient pores, that water molecules occasionally traversed, were observed. (e) Quantification of the pore diameter, at a single point in the simulation, along the length of the twelve peptide Ct21 pore passing though the bilayer. Zero on the x-axis indicates the midpoint of the toroidal pore. A representative twelve peptide pore is also shown. (f) Quantification of the smallest diameter of the twelve peptide Ct21 pore throughout the 500 ns simulation. (g) Quantification of pore diameter from MD simulations with reducing Ct21 peptide number. A twelve-peptide pore was initially formed, followed by removal of single peptides, stepwise, to examine how reducing peptide number affects pore properties. The average pore diameter from 12, 11, 10, 9 and 4 peptides are 2.6 ± 0.3, 1.6 ± 0.3, 1.1 ± 0.5, 1.1 ± 0.4 and 1.0 ± 0.3 nm, respectively. The box and whisker plot was generated with 5th (min) and 95th (max) percentiles and an interquartile range representing the 25th and 75th percentiles. The center line represents the median.
