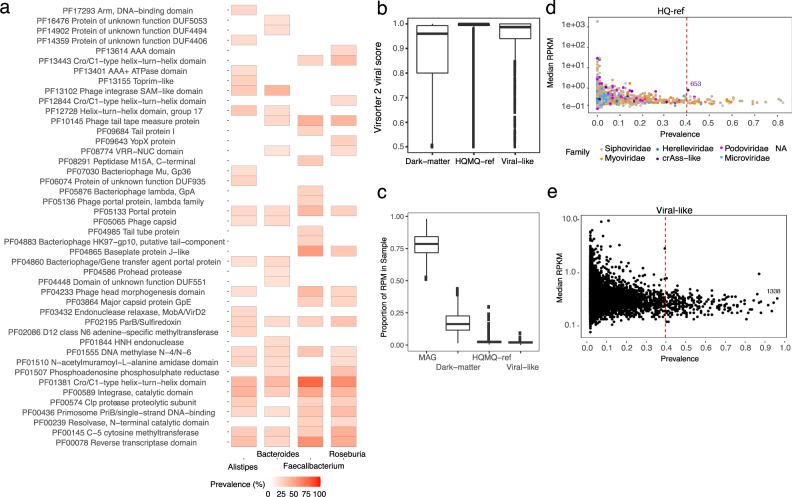
Fig. 6. Viral proteins and the dark-matter metavirome.
a The percentage of HQ viruses, associated with four bacterial host genera; Alistipes, Bacteroides, Faecalibacterium and Roseburia, which encode top-20 prevalent PFAM domains. b Virsorter2 viral prediction scores for all viral bins with at least one viral hallmark gene. Completeness was estimated using CheckV and the bins were grouped as (1) HQ-MQ-ref when completeness ≥50% or high-quality ≥90% (n = 45,983 bins), (2) bins with less than 50% completeness were annotated as Dark-matter (n = 392,226 bins), and (3) dark-matter bins with confident CRISPR spacers against a bacterial host were annotated as Viral-like (n = 43,695 bins). c The distribution of sample RPM of bacterial MAGs, HQ-MQ-ref viral populations, Dark-matter and Viral-like populations as defined in (b). The majority of sample reads were mapped to MAGs but on average 17.7% of all reads mapped to Dark-matter bins. d The abundance in RPKM of rare and highly prevalent viruses with an HQ genome in HMP2. Each point represents a viral population coloured according to the viral taxonomic family. The progenitor-crAssphage is indicated as cluster 653. e As in (d) but with viral-like populations like cluster 1338 showing that many are low abundant, but highly prevalent. RPM read per million, RPKM read per kilobase million.