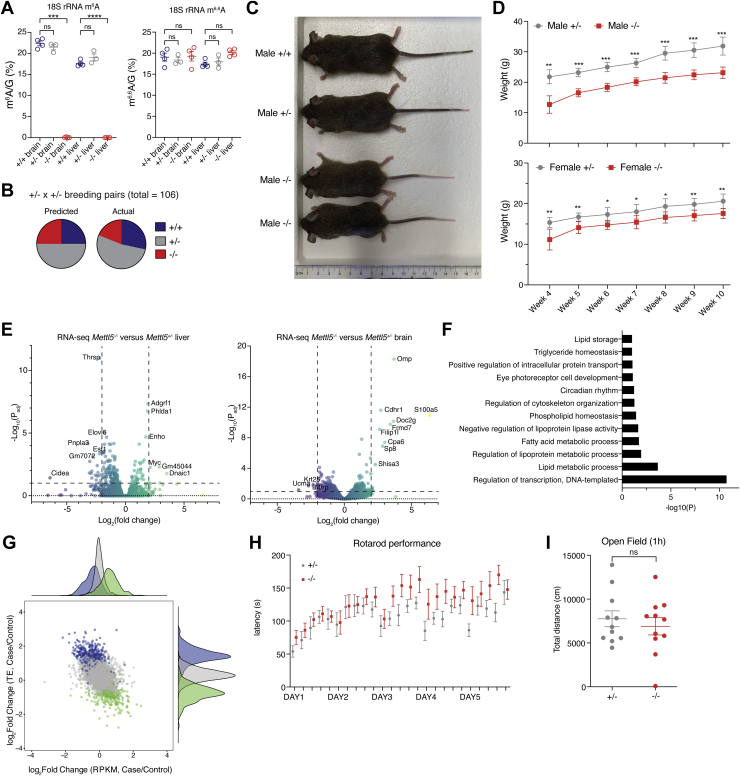
Figure 4.
Mettl5 knockout mice have developmental and metabolic defects.A, levels of m6A (left) and m6,6A (right), normalized to G, obtained by LC-MS/MS of 40-nt probe-purified segments of 18S rRNA surrounding the m6A 1832 site from the brains and livers of Mettl5+/+, Mettl5+/−, and Mettl5−/− mice. n = 4, Mettl5+/+, n = 3, Mettl5+/−, and n = 4, Mettl5−/−. Analyzed by one-way ANOVA comparison with Sidak test for multiple comparisons. B, pie chart of the predicted (left) and actual (right) mice born of wild type (+/+, dark blue), heterozygous (+/−, grey), and knockout (−/−, red) genotypes from heterozygous x heterozygous breeding pairs (total = 106 mice; 30 +/+, 56 +/−, 20 −/− mice). C, photograph of male littermate Mettl5+/+, Mettl5+/−, and Mettl5−/− mice at 8 weeks of age. D, body weights in grams of male (top) and female (bottom) Mettl5+/− and Mettl5−/− mice from 4 weeks (weaning) to 10 weeks of age. n = 6 mice for male +/−, n = 4 for male −/−, n = 5 for female +/− and −/−; unpaired t test was used to compare each time point. E, volcano plots from mouse liver (left) and brain (right) RNA-seq data displaying the negative log10 of the adjusted p value versus the log2 fold change comparing the transcriptomes of Mettl5−/− and Mettl5+/− mice (n = 3 pairs). F, dysregulated pathways based on analysis of RNA-seq data from Mettl5−/− and Mettl5+/− mouse livers by the software DAVID (65). G, effects of METTL5 knockout on translation and transcription as displayed by log2(fold change) of translation efficiency (TE) versus log2(fold change) of reads per kilobase of transcript per million mapped reads (RPKM) from ribosome profiling input and ribosome-protected fragment sequencing of mouse livers. Case = Mettl5−/−, Control = Mettl5+/+. H, latency in seconds for Mettl5+/− and Mettl5−/− mice to fall off the rotarod in a rotarod performance test. I, total distance in centimeters travelled by Mettl5+/− and Mettl5−/− mice when placed in a new environment in an open field test. Unpaired t test shown. H and I, n = 11 pairs. ns: not significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005.