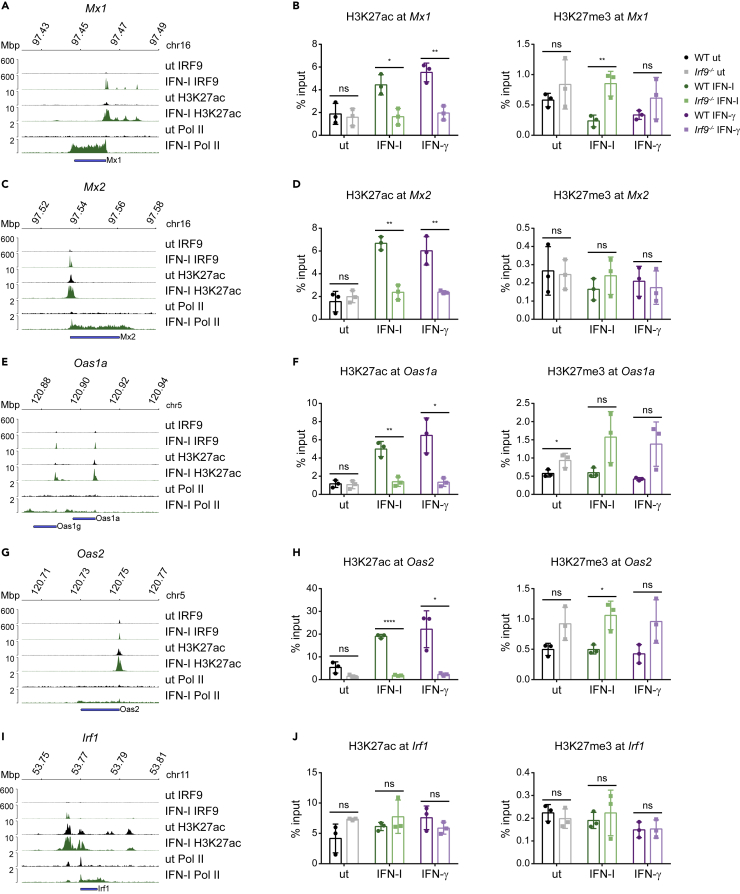
Figure 7.
Histone modifications at ISG promoters correlate with the local chromatin accessibility
(A, C, E, G, and I) Genome browser tracks representing published ChIP-seq datasets in BMDM with antibodies against the following proteins after IFN-I stimulation: IRF9 (1.5h), H3K27ac (1h) and Pol II (2h) showing promoter occupancy at indicated gene loci (one representative replicate).
(B, D, F, H, and J) Site-directed ChIPs (Mx1, Mx2, Oas1a, Oas2, and Irf1) were performed in bone marrow-derived macrophages isolated from wildtype and Irf9−/− BMDM, treated for 2h either with IFN-I or IFN-γ and processed for ChIP with antibodies targeting H3K27ac and H3K27me3 as indicated (See also Figure S5). Data represent mean ± SD of biological triplicates, analyzed for statistical significance by Student’s two-tailed, unpaired t-test using Graph Pad Prism software. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001