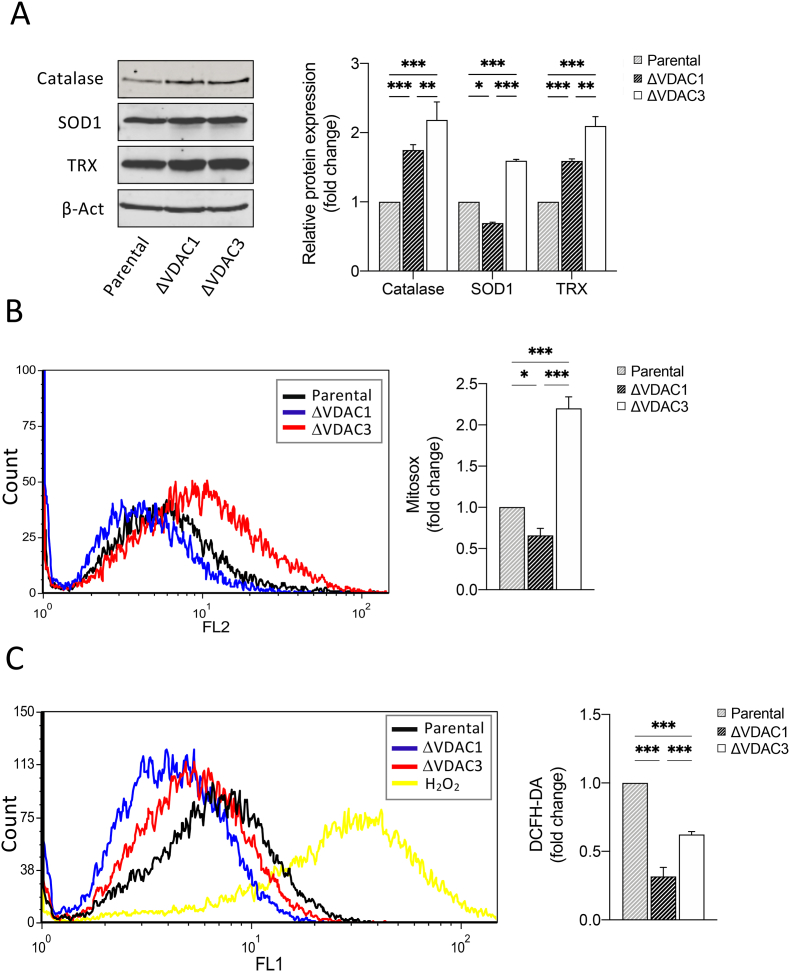
Fig. 2.
Analysis of mitochondrial superoxide content and quantification of the expression levels of the main antioxidant enzymes in HAP1 cell lines. A. Representative immunoblot and relative protein quantification of antioxidant enzymes (Catalase, superoxide dismutase 1 (SOD1) and Thioredoxin (TRX)) in HAP1-ΔVDAC1 and HAP1-ΔVDAC3. Data are normalized to the β-actin, expressed as means ± SEM (n = 3) and compared to HAP1 parental cell line. B. Mitochondrial superoxide determination by MitoSOX red probe. Representative MitoSOX Red fluorescence profiles evaluated by flow cytometry in each population of HAP1 cell lines (left). Quantitative analysis of the mean fluorescence intensity MitoSOX Red in each population of HAP1 cell line. Results are expressed as the mean ± SD (n = 3) and compared to HAP1 parental cells (right). Results are expressed as the mean ± SD (n = 3) and compared to HAP1 parental cells. C. Intracellular ROS detected by DCFH-DA fluorescence measured by flow cytometer. Representative fluorescence profiles detected by flow cytometry for each cell line (left). Quantitative analysis of the average fluorescence intensity of DCFH-DA (%) in each gated population of HAP1 cell lines (right). Results are expressed as the mean ± SD (n = 3) and compared to HAP1 parental cell line. Data were analyzed with Two-Way ANOVA or One-Way ANOVA; with *p < 0.05 **p < 0.01 and ***p < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)