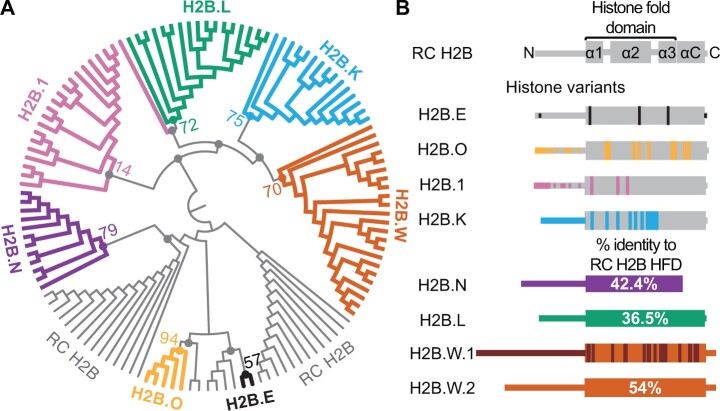
Fig. 1.
Phylogenomic analyses identify distinct H2B variant clades in mammals. (A) A maximum-likelihood protein phylogeny of the HFD of selected ancestral/RC H2B sequences and all intact H2B variants sequences from 18 representative mammalian species is represented as a circular cladogram (see supplementary data 1, Supplementary Material online, for a phylogram with branch lengths scaled to divergence). RC H2B histones are shown in gray, and seven H2B variant clades identified using phylogeny are highlighted in colors: H2B.E (black), H2B.O (yellow), H2B.N (purple), H2B.1 (pink), H2B.L (green), H2B.K (blue), and H2B.W (orange). Bootstrap values at selected nodes with >50% support are shown along with colored dots to indicate the nodes they represent. The H2B.1 clade has a low bootstrap support of 14% owing to its high similarity to RC H2B (see supplementary figs. S1 and S3, Supplementary Material online, for additional information). Select nodes with low bootstrap support values (<20%) are indicated with a gray dot. (B) Schematics of RC H2B and H2B variants. A structural schematic of a RC H2B at the top shows the N-terminus, HFD (including the α1, α2, α3 helices, and intervening loops), αC domain, and the C-terminus. Variants with high identity to RC H2B (H2B.E, H2B.O, H2B.1, and H2B.K) are shown in gray with differences from RC H2B colored using the same colors as (A). More divergent variants (H2B.N, H2B.L, and H2B.W.1 and H2B.W.2) are represented in solid colors and the percent identities of the HFD and αC domain compared with RC H2B are indicated. Differences between H2B.W.1 and H2B.W.2 are further highlighted in brown to indicate the divergence of these paralogs. Schematics and percent identities are based on human sequences, except for H2B.E and H2B.O, which are only found in some rodents (mouse sequence used) and platypus, respectively, and H2B.L which is pseudogenized in humans, (rhesus macaque sequence used as a reference).