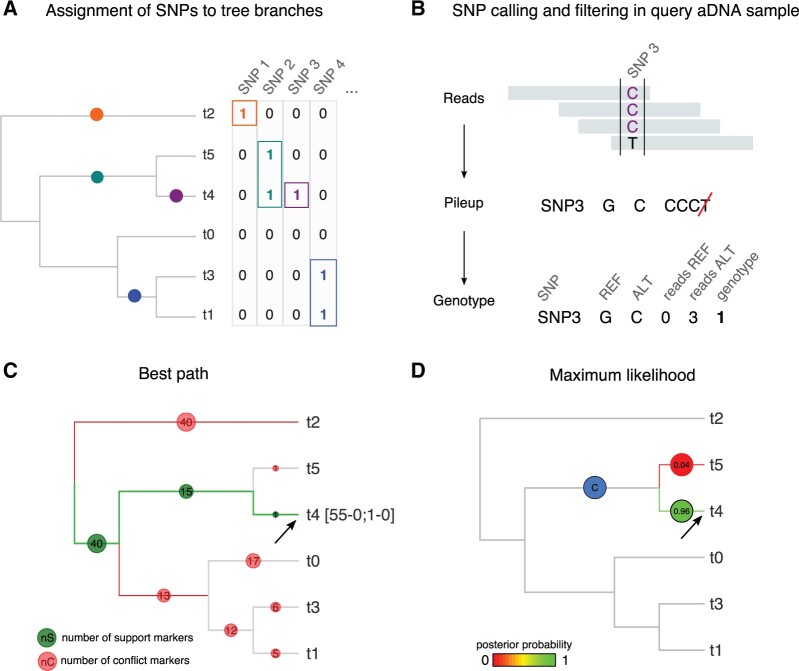
Fig. 1.
Overview of pathPhynder workflow. We illustrate the method using a small simulated data set of 6 reference samples and 112 SNPs. (A) The initial step is the assignment of phylogenetically informative SNPs in the reference data set to branches. This can be achieved with phynder by estimating the likelihood of each SNP at any given branch of the tree. (B) A pileup from aDNA reads is generated at each SNP, then filtered for mismatches and potential deamination. Here, because SNP3 is defined by alleles G and C, the T base is excluded as likely to be caused by post-mortem deamination. (C) Best path method: aDNA sample genotypes for each SNP are assigned to the corresponding branch of the tree and binned into support and conflict categories. In this case the best path is supported by 56 derived markers (green), of which 55 are above the assigned branch and one is on the branch, with no conflicting markers along the chosen path [55-0; 1-0]. (D) Maximum likelihood method: the likelihoods for placing the query sample on each edge of the tree are converted to posterior probabilities using Bayes’ rule and branches with posterior probability greater than 0.01 are indicated (largest posterior in green). The blue circle shows the lowest branch in the tree for which the sum of posterior probabilities for the whole clade below that branch (including the branch in question) is greater than 0.99, providing a conservative assignment when placement is uncertain. The arrows point to the correct location for the query sample.