Abstract
Genetically encoded reporters have greatly increased our understanding of biology. While fluorescent reporters have been widely used, photostability and phototoxicity have hindered their use in long-term experiments. Bioluminescence overcomes some of these challenges but requires the addition of an exogenous luciferin limiting its use. Using a modular approach we have engineered Autonomous Molecular BioluminEscent Reporter (AMBER), an indicator of membrane potential. Unlike other luciferase-luciferin bioluminescent indicators, AMBER encodes the genes to express both the luciferase and luciferin. AMBER is a voltage-gated luciferase coupling the functionalities of the Ciona voltage-sensing domain (VSD) and bacterial luciferase, luxAB. When AMBER is co-expressed with the luciferin producing genes it reversibly switches the bioluminescent intensity as a function of membrane potential. Using biophysical and biochemical methods we show that AMBER switches its enzymatic activity from an OFF to ON state as a function of the membrane potential. Upon depolarization AMBER switches from a low enzymatic activity state to a high enzymatic activity state, showing several-fold increase in the luminescent (ΔL/L) output. We expressed AMBER in the pharyngeal muscles and mechanosensory neurons of C. elegans. Using the bioluminescent voltage signals and compressed sensing techniques we reconstructed the electropharngeogram of the C. elegans pharynx, validating the sensor function in vivo. Thus, AMBER represents the first fully genetically-encoded bioluminescent reporter, without requiring exogenous luciferin addition.
Keywords: Bioluminescence, Voltage Imaging, Bacterial Luciferase
Graphical Abstract
1. Introduction
Genetically-encoded optical sensors have gained prominence for reporting protein-protein interactions, gene expression, and cellular signaling. Most reporters provide an optical readout of the signal of interest via the excitation of a fluorescent protein [1]. Application of molecular engineering to detect, record and modulate neural signatures is a major area underpinning several neurotechnology initiatives. Over the last decade, there has been exponential growth in the development of genetically engineered molecular cellular probes owing to their ability to precisely target tissues of interest. A wide range of biomolecular sensors currently exists for detecting changes in voltage [2, 3], and the levels of calcium [2, 4, 5], potassium [6, 7]and neurotransmitters[8, 9, 10]. While fluorescent reporters have been the mainstay, they suffer from photobleaching and phototoxicity affecting their long-term use [11]. Near-infrared indicators [4, 12, 13] and two-photon excitation [14, 15] overcome some of these limitations [16, 17], but the development of efficient indicators of cellular activity remains an issue.
Intracellular calcium (Ca2+) concentration and membrane potential are the two well characterized proxies of neuronal activity. Several Genetically-Encoded Ca2+ Indicators (GECIs) [2, 4, 5, 18] were developed over the last two decades with GCaMP variants [19] being most widely employed in the neuroscience community. However, the relationship between Ca2+ signals and neuronal activity can vary among different neuronal types and may become completely uncoupled [20, 21]. Despite the great utility of GECIs, they have additional limitations. For instance, several organelles (endoplasmic reticulum and mitochondria) and regulatory proteins contribute to Ca2+ compartmentalization [22, 23]in neurons. Over expression of GECIs in neurons can cause these Ca2+-responsive probes to contribute to Ca2+ buffering, thereby affecting neurophysiology [23]. Despite its high Signal-to-Noise ratio (SNR), GECIs cannot report subthreshold or hyperpolarized potentials [24, 25]as is possible with fluorescent Genetically Encoded Voltage Indicators (GEVIs) [26]. Recently, a fluorescent hybrid GEVI complexing with an exogenously supplied synthetic dye was reported to be stable while recording for tens of minutes[27]. However, deciphering the information processing in circuits with a large population of neurons necessitates conducting experiments for extended periods, demanding molecular voltage reporters with exceptional photostability. Apart from its ubiquitous role in the action potential, the dynamic membrane potential is emerging as a critical factor in controlling many crucial cellular processes, including cell cycle progression, proliferation, and wound healing[28, 29]. Elucidating this emerging role of the membrane potential requires new tools and methods for actively recording the membrane potential over long periods (days).
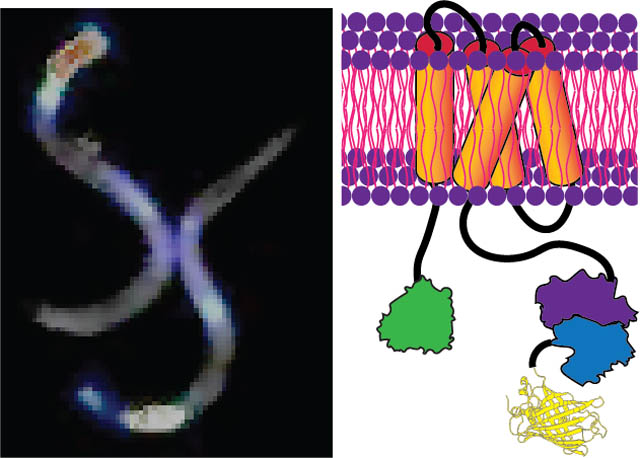
Recently bioluminescence has garnered attention as an alternative to overcome some of the limitations of fluorescent GEVIs and GECIs [30], especially its ability to record for prolonged periods of time. Bioluminescence imaging does not require any external illumination, and light emission is achieved as a by-product of a biochemical reaction – oxidation of a substrate (luciferin) catalyzed by an enzyme (luciferase). Commonly used light-generating luciferases from fireflies (Lampyridae) and marine organisms such as Aequoria victoria, Renilla reniformis, Gaussia princeps, Metridia longa, and Vargula hilgendorfii [31, 32, 33, 34, 35] require exogenous addition of luciferin since the biosynthetic pathways for in situ luciferin production have not been identified in most organisms. There are two exceptions - the genes coding for the fungal luciferin, 3-hydroxyhispidin [36] and the bacterial luciferin, a molecular complex of fatty aldehyde and reduced flavin mononucleotide, FMNH2 [37]. In a tour-de-force, Kotlobay et al. [38] discovered a set of genes required for expressing both the fungal luciferase and luciferin. However, the fungal bioluminescence system requires a total of seven genes assembled from multiple organisms, which can be challenging to express efficiently in eukaryotic systems. In contrast, the genes encoded in the bacterial lux operon are in a single polycistronic mRNA [39] and can enable enhanced bioluminescence [40]. The lux operon, unlike other bioluminescent systems, consists of a series of six genes (luxCDABE and the flavin oxidoreductase gene, frp) synergistically combining to produce both the luciferase and a luciferin-generating secondary protein complex (Figure S1 in the Supporting Information, SI). The luciferin synthesizing protein complex recycles the products (myristic acid and FMN) and the intermediates of the light reaction endogenously to light generating substrates (myristyl aldehyde, also known as tetradecanal and FMNH2) using metabolic pathways [40]. Fortuitously, tetradecanal is not freely available in large quantities in eukaryotes [41], ensuring low background activation. There were concerns about apparent cytotoxic effects of aldehyde compounds in eukaryotes [42] but the concentration of tetradecanal synthesized using lux operon expression does not seem to attain the toxic dose affecting cell viability [41]. Recent work also confirmed there is no cytotoxic effect when the lux cassette is expressed in mammalian cells [43]. In this paper, we describe a new type of Luminescent Voltage Indicator (LuVI) named AMBER (Autonomous Molecular BioluminEscent Reporter) for detecting voltage activity in freely moving animals. AMBER uses the functionalities of the ascidian Ciona Intestinalis voltage sensing domain (VSD) [44], a synthetic enhanced bacterial luciferase (derived from P. luminescens luxAB, pl-luxAB) [41], and a fluorescent moiety, YPet [45]. AMBER also leverages the molecular architecture of a previously developed GEVI, VSFP2.1 [46]. The rationale behind this choice stems from the efficient coupling between the conformational change of the VSD and the Cerulean/Citrine FRET (Förster Resonance Energy Transfer) pair of VSFP2.1 while operating within the physiological voltage range. While VSFP2.1 responds by modulating the FRET distance, AMBER provides an increase in overall activity by switching from an inactive to an active state upon depolarization. AMBER provides an unprecedentedly large dynamic range in the optical readout and an increased emission efficiency. Unlike other voltage probes reported to date, AMBER switches from an OFF to ON state upon depolarization. The overall light emission is ≈ 3 times brighter than the initial resting signal, and an on-off ratio greater than LOTUS-V [47, 48], the first LuVI developed using the deep shrimp luciferase [49]. We demonstrated in vivo functionality by expressing AMBER in C. elegans pharyngeal muscles and mechanosensory neurons.
2. Results
2.1. Optimizing Engineered Bioluminescent Voltage Indicators
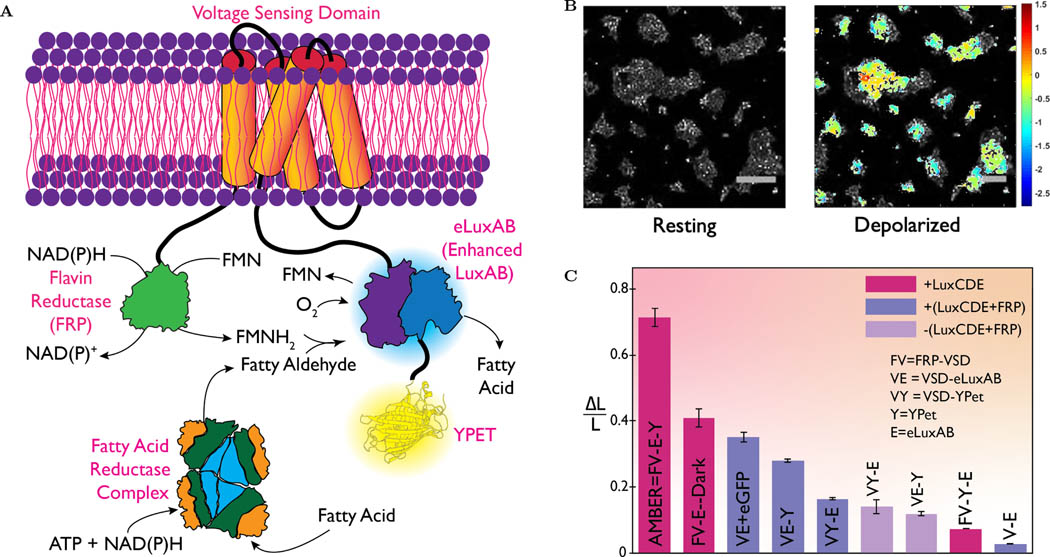
We postulated voltage sensing ability can be conferred to the lux-based LuVIs by replacing the fluorescent FRET donor, Cerulean, of VSFP2.1 [46] with the mammalian codon-optimized synthetic luciferase, enhanced luxAB (‘eluxAB’) [50] and the FRET acceptor, Citrine with YPet. This replacement resulted in the protein construct VSD-eluxAB-YPet (referred to as VE-Y). For the sake of brevity all engineered protein constructs are named using the first letter of each of the functional domains (V- Voltage sensor, E- eluxAB, Y- YPet, and F- FRP). Previous reports on Bioluminescence Resonant Energy Transfer (BRET) performance show that the acceptor position could influence the brightness of the probe [51]. Therefore, we swapped the position of eluxAB and YPet in VE-Y to compare its performance with the VY-E (see Figure S2). Molecular cloning approaches to create plasmids with relevant genetic elements for expressing various protein constructs are described separately (see the SI section 1.2 and Table S1). Production of FMNH2 is the rate-limiting step in the bacterial bioluminescence reaction [52]. A direct transfer of FMNH2 minimizes FMNH2 oxidation via the dark pathway, thereby significantly increasing the bioluminescence quantum yield[53, 54]. We wondered if placing FRP in close apposition to eluxAB could enable direct transfer of FMNH2 to eluxAB from the FRP-FMNH2 complex and thereby increase the bioluminescent intensity. Therefore, we fused the FRP domain to the N-termini of both VE-Y and VY-E to obtain FV-E-Y and FV-Y-E, respectively (See the SI Section 1.2). We did not evaluate the positioning of FRP at the C-terminus since it is closer to the putative voltage activated S4 segment and VSD switching could become excessively slow if large protein domains are fused to its C-terminus. Furthermore, we wanted to mimic the overall structure of the voltage gated phosphatase and needed a terminal β-barrel structure (provided by YPet).
Plasmid DNAs encoding the four principal engineered protein constructs - VE-Y, VY-E, FV-E-Y, and FV-Y-E were co-expressed with their substrate producing proteins (FRP-luxCDE for VE-Y and VY-E and luxCDE for FV-Y-E and FV-E-Y) in HEK293 cells (See the SI Section 1.3). Bioluminescent images were recorded using a custom-built imaging set up (See the SI Section 1.4) placed inside a dark room. The recorded images were subsequently processed using the ImageJ software (See the SI Section 1.12). The efficacy of all engineered constructs was tested in mammalian cells at resting and depolarization states (using ≈ 50mM KCl). Brightfield and bioluminescent images of cells expressing different protein constructs were obtained before and after KCl addition (Figure S2). VE-Y performed better than VYE in modulating the eluxAB activity. Surprisingly FV-Y-E performed the poorest of all the constructs tested, suggesting the positioning of eluxAB and YPet affects the light modulation more than the proximity to FRP. However, as expected FV-E-Y performed the best and this result confirms that placing eluxAB and FRP domains in close apposition increases the propensity of direct FMNH2 transfer from FRP to eluxAB. Figure 1A shows a schematic of FV-E-Y (henceforth referred to as ‘AMBER’) that exhibits the highest change in bioluminescent signal compared to other constructs. Figure 1B shows an overlay of color-coded contour pixels quantifying the fractional luminescence (ΔL/L) on the corresponding brightfield image of HEK293 cells expressing AMBER.
Figure 1:
Molecular engineering to maximize autobioluminescence brightness. (A) schematic of the membrane resident AMBER probe is engineered as a fused polypeptide chain, NH2-FRP-VSD-eluxAB-YPet-COOH. Light reaction is catalyzed by eluxAB domain, which requires fatty aldehyde (produced by the cytosolic Fatty Acid Reductase complex consuming ATP), FMNH2 (produced by FRP consuming NAD(P)H) and molecular oxygen. Placing FRP in close apposition to eluxAB decreases the chances of FMNH2 auto oxidation either by direct transfer from FRP to eluxAB or by rapid diffusion. When VSD undergoes conformational change upon membrane depolarization, the beta barrel of the YPet turns on the enzymatic activity of eluxAB producing a several fold-increased bioluminescence (λmax ≈ 490nm.)(B) A contour of fractional luminescence (log10ΔL/L) of AMBER superimposed over the corresponding brightfield image of HEK293 cells before and KCl addition. AMBER shows 28-fold maximal increase and 72% average increase from 85% of chosen population of cells. The scale bar length is 200μm. (C) A plot of normalized fractional luminescence confirms that AMBER performs best among all the engineered protein constructs followed by the Dark Mutant. Plotted data represents mean ± std. error of the pixel recordings. (Wilcoxon signed-Rank test; n > 25 cells for all constructs; p < 10−5)
We formulated a new metric termed ‘Normalized Fractional Luminescence’ to quantitatively compare the performance across constructs. Normalized fractional luminescence is obtained by normalizing the mean photon flux (average differential intensity per unit exposure time) of a chosen construct by the maximum achievable value among all constructs (i.e. the photon-flux ratio of AMBER, see the SI Section 1.12). Figure 1C shows an unbiased comparison of the bioluminescent output for all the engineered constructs. FV-E-Dark ranks second and its structure was the same as AMBER except for the mutated YPet chromophore to abrogate its fluorescent property. This was followed by a set of candidates whose N-termini did not contain the FRP domain, illustrating the role of FRP in improving the brightness. Light due to endogenously produced substrates (Figure S4) was very dim – about 10 times smaller than the maximum achieved signal even though the integration time was three times as long. In contrast, un-transfected cells showed no detectable intensity increase after KCl addition (Figure S3). Candidate rankings made based on normalized fractional luminescence agrees with the outcomes of statistical approach using information entropy (See the SI Section 1.15 and Table S4). We performed Western blot assay to ensure the expression of the engineered proteins are at the comparable levels (See the SI Section 1.16 and Figure S19)
2.2. Emission Spectrum of AMBER
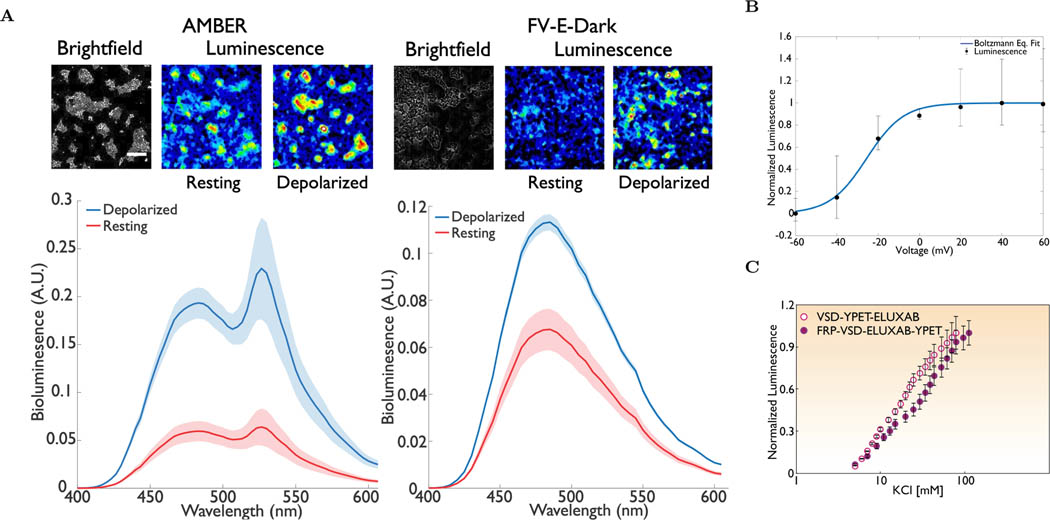
An increase in the bioluminescence by several-fold after KCl addition precludes BRET from being the primary contributor. We therefore performed a more quantitative assay by recording the emission spectra using a plate reader (see the SI Section 1.5). The bioluminescent spectra of HEK293 cells expressing AMBER were recorded before and after KCl addition (See Figure 2A(left)). Representative bioluminescent micrographs obtained from imaging experiments are also included to enable visualization. We applied a weighted-residual least square approach to curve fit a non-linear regression model through recorded spectral intensities (See the SI Section 1.5 and Table S2). A spectral curve fitting model was represented as a linear weighted combination of three a priori inputs - background bioluminescence of HEK293 cells suspended in the growth media recorded separately, the bioluminescence emission spectrum of iluxAB (derived from pl-luxAB) published previously [40] and the fluorescence emission spectrum of YPet[55]. The shaded region along each trace represents the 95% confidence interval. Spectral intensities at the eluxAB donor (≈ 490nm) and the YPet acceptor (≈ 530nm) wavelengths increased after KCl addition approximately by the same factor (≈ 3X). This result suggests that conformational change of the VSD upon membrane depolarization does not seem to alter the Förster distance between eluxAB and YPet. Therefore, BRET is not the major contributor to increased light emission after KCl addition.
Figure 2:
Spectral emission and voltage response of AMBER. (A, left) Representative micrographs (top) and the bioluminescent emission spectrum (bottom) of AMBER at the resting and KCl induced depolarization states (Wilcoxon signed-Rank test; n > 25 cells; p <10−5). A ≈ 3-fold increase in the average signal was observed after KCl addition. There was negligible difference in the ratio of the donor (λex=490nm) to acceptor (λem=530nm) signals before and after KCl addition. Shaded region represents 95% confidence interval. (A, Right) Representative micrographs (top) and the bioluminescent emission spectrum (bottom) of FV-E-Dark at the resting and KCl induced depolarization states. Beta barrel of the Dark mutant is sufficient to switch the activity of eluxAB domain. The loss of BRET component is evident from the decreased brightness of the micrographs. Shaded region represents 95% confidence interval. (B) Representative whole cell voltage clamp recording of a HEK293 cell expressing AMBER. Recordings from six independent repeats varying the membrane potential between −60mV and 60mV at an increment of 20mV were plotted. Electrically stimulated optical readout follows the classical Boltzmann distribution with V1/2 ≈ −25mV. The Boltzmann equation (1/(1 + exp(−q(V − V1/2)/zQkBT))) fit yields a value of V1/2 = −25.5 ± 1mV and a zQ ≈ 3 ± 0.25. A single tailed paired t-test statistic confirms statistically significant difference between the mean intensities corresponding to the off and on states (Shapiro Wilk normality p-value ≈ 0.562 > Significance level, α = 0.05, single tailed paired t-test p < 0.02). AMBER shows approximately ≈3X increase in luminescence in experiments performed on HEK293T. (C) Normalized bioluminescent response of the AMBER and VY-E under KCl titration. An order of magnitude change in the extracellular KCl concentration is necessary to achieve the maximum intensity for the chosen constructs. For both the patch clamp and KCl depolarization experiments the luminescence is the total light collected over all wavelengths. Scale bar length is 200μm.
2.3. Performance of the Dark Mutant
Additional evidence to confirm the switching of AMBER upon depolarization was obtained by mutating residues GLY65THR and GLY67ALA within the chromophore of YPet in FV-E-Y[56] (See the SI Section 1.2). These mutations (creating the construct FV-E-Dark termed as the ‘Dark mutant’), almost completely eliminate BRET. Figure 2A(right) shows the bioluminescent emission spectra of HEK293 cells expressing FV-E-Dark construct at the resting and depolarized states. Representative bioluminescent micrographs were also included. The spectral curve-fitting model for the Dark mutant was represented as a linear weighted combination of two of the aforementioned a priori inputs – the background bioluminescence and the iluxAB emission spectra (See the SI Section 1.5 and Table S2). Bioluminescent emission of HEK293 cells expressing FV-E-Y-Dark increased significantly after KCl addition as expected. An increase in the spectral intensity at ≈ 490 nm by a factor of ≈ 2× upon depolarization suggests that switching of the enzymatic activity as the dominant mode of the light emission. However, the BRET mechanism seems to increase the overall emission and thus AMBER emission is brighter than FV-E-Dark
2.4. NADH Assay
Tracking NADH production provides an indirect evidence for eluxAB enzymatic activity since NADH production increases with the drop in cellular O2 concentration. NADH is utilized both for regenerating FMNH2 from FMN catalyzed by FRP and for the fatty aldehyde synthesis from the fatty acid via the luxCDE complex. While the reduced form (NADH) is fluorescent, the oxidized form (NAD+) is not. We monitored endogenous NADH fluorescence before and after KCl addition, which showed an increase in the NADH fluorescence after KCl addition (See the SI Section 1.4). While the steady-state NADH fluorescence before KCl addition is not large enough to be detected, a step increase after KCl addition invariably points to the drop in O2 concentration. Cytosolic NADH is produced at detectable levels within a few seconds after O2 consumption during the light reaction (Figure S5), and similar observations have been reported earlier [57]. Thus, we confirmed the causal link between depolarization and bioluminescence-induced O2 consumption, as evidenced by the increase in the NADH fluorescence. This strongly supports a voltage-gated enzymatic switching mechanism. An increase in NADH fluorescence also indicates that the rate of NADH production due to an O2 drop is faster than the rate of NADH consumption for aldehyde synthesis. This suggests that steady-state aldehyde concentration in cells produced by luxCDE is sufficient to carry out the light reaction for a long duration (at least for a few tens of minutes),
2.5. Voltage Switching Characterization
We performed electrophysiology experiments to determine the voltage-dependent characteristics of AMBER. We transiently co-expressed AMBER and luxCDE in HEK293 cells to record single-cell bioluminescence under voltage clamp. Results from the patch-clamp experiments are shown in Figure 2B (See the SI Section 1.6). The curve follows the classical Boltzmann activation and the potential where the activity reaches half the maximal value termed V1/2 is the point of maximal sensitivity. The half-maximal voltage of AMBER, V1/2 ≈−25 mV is within the physiological range of neuronal action potentials similar to other GEVIs [58, 59, 60]. In comparison, the V1/2 of the bioluminescent voltage indicator LOTUSV [47] is ≈ 0mV. Ionic depolarization of HEK293 cells expressing AMBER and VY-E (with luxCDEFRP) was done by titrating with incremental amounts of KCl (Figure 2C). The V1/2 deduced applying Nernstian relation to the KCl titration data agrees closely with the value obtained via patch-clamp recordings. Interestingly, V1/2 of AMBER is more positive compared to that of VSFP2.1 (V1/2 ≈−70 mV) but agrees closely with the switching threshold of VSFP2.3, derived from VSFP2.1 by modifying the linker length between the donor and the VSD. Additionally, V1/2 shifts towards the positive direction for AMBER compared to VY-E. We do not know if the BRET pair interacts with the VSD domain and whether its rearrangement caused the voltage shift. Earlier work showed evidence for an interaction between the fluorescent protein and the VSD domain, but the mechanism of the fluorescence modulation by the VSD is not fully understood[58]. We speculate that the interaction between the YPet and the VSD domains could account for the observed shift.
2.6. Chemogenetic Activation
An increase in extracellular K+ promotes depolarization, thereby affecting many cellular signaling processes. We therefore wanted to independently verify if enzymatic switching could be achieved using an activator other than K+ under physiological conditions. We achieved this by co-expressing rat capsaicin receptor (rTRPV1) with the AMBER (and its substrate luxCDE) in HEK293 cells and stimulated the cells using capsaicin [61] (See the SI Section 1.4). We chose rTRPV1 because it has a large single-channel conductance and capsaicin-induced activation of rTRPV1 causes cellular depolarization due to the large inward cation current [62]. Bioluminescence experiments performed under these conditions also exhibited changes in bioluminescence similar to depolarization with K+ (Figure S6 in the SI)
2.7. Characterization of AMBER Kinetics
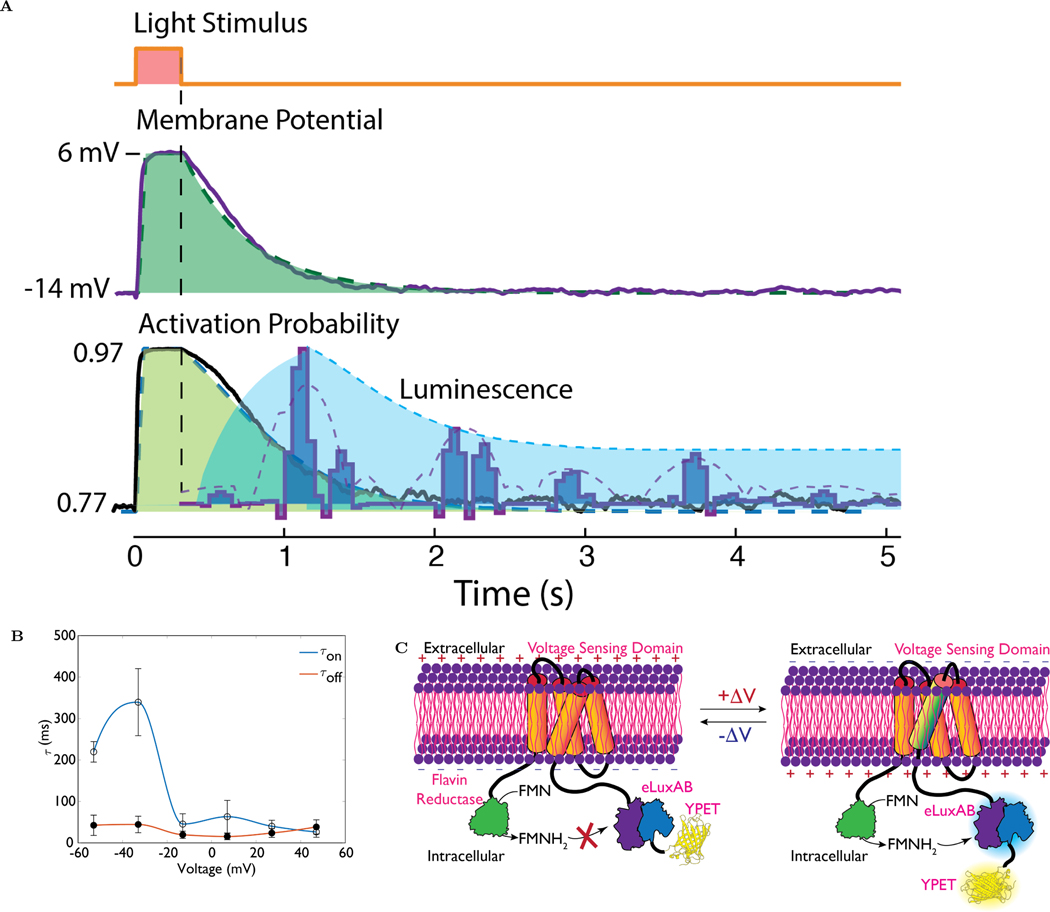
We characterized the AMBER kinetics using a custom-built all-optical electrophysiology rig. Briefly, AMBER was co-expressed with a red light-sensitive optogenetic probe, ReaCHR [63]in HEK293 cells. Bioluminescence was recorded following cellular depolarization for 300ms using a red light (λ=620 nm). We consistently observed that the resting potential of cells co-expressing AMBER and ReaCHR (≈−14mV) was more positive than a cell expressing ReaCHR alone (≈ −25mV) possibly due to AMBER induced ReaCHR activation under resting conditions (see Figure S12 in the SI). We accounted for this variability in the resting state of a cell while quantifying the voltage transients using the Boltzmann statistics shown in Figure 2B. We note the slow decay of the voltage over time allows applying quasi-equilibrium approximation of the voltage trace so that the Boltzmann statistics can be used in these kinetic conditions. Figure 3A shows the signals recorded in the all-optical electrophysiology experiment. The top trace shows the onset and deactivation of a 300ms light pulse. The second trace shows the membrane potential of a cell co-expressing AMBER and ReaCHR and the best fit single exponential decay (τoff = 450ms) is shown as an overlay. The third trace shows the computed activation probability using the Boltzmann statistics illustrating the cell is in a fairly depolarized state even in the absence of the light stimulus and thus has a considerable baseline activity. The change in activity is 20% under light stimulation in these experiments. In a separate experiment we recorded the bioluminescent signal under identical optical stimulation conditions and the resultant bioluminescent signals are overlaid onto the activation probability trace. The signals were collected for 8 stimulation pulses repeated at a 5s interval. The data shown is the median of the data collected from 8 cells (See Figure S13 for additional intensity traces). We noticed a slow frequency envelope (purple dotted line in figure 3A) on the bioluminescent signals and attribute this to the depletion of the substrate and its subsequent regeneration. The spacing between the pulses is between ≈ 0.7–1 second, the burst themselves having an activation time constant of ≈ 300ms and deactivation time constant of ≈ 200ms. Since the time constant of the charge movement of the VSD is known to correspond to the fast component of fluorescent GEVIs [64], we characterized the charge activation and deactivation time constants,τon and τoff respectively, of cells expressing AMBER and the results are shown in figure 3B(see SI section 1.6). The τoff of AMBER (15–45 ms) is slightly faster than the deactivation time constant of the parent VSFP2.1 (60–80 ms)[46]. On the other hand, the τon of AMBER (≈26–330 ms) has a considerably larger variation than VSFP2.1 (≈10–85 ms). Beyond the V1/2 of AMBER the on-time constants are more closely matched (26–63 ms) to VSFP2.1. Thus the introduction of the FRP, eluxAB and YPet has a pronounced effect on the activation time constant when the cell is in a hyperpolarized state. Taken together the slow component of the time constant of AMBER deduced from the slow envelope function is expected to be in the order of ≈200–300 ms and the fast time constant is the order of ≈25–50ms as given by the charge activation and deactivation time constants. In comparison, LOTUS-V exhibits a fast and slow time constant, with 30–40% of the population in the fast population. The fast time constants are 3.09 ms for the activation and 6.12 ms for the deactivation, while the slow component was 204ms and 144ms for the activation and deactivation respectively.
Figure 3:
(A)All Optical Electrophysiology to study the kinetis of AMBER. HEK293 cells containing a channel rhodopsin (ReaChr[63] is activated by a brief 300ms 620 nm light pulse (top trace). The resulting membrane potential is shown in the middle trace, the shaded green region is the best fitting single exponential decay to the voltage curve. The decay time constant is 450 ms. The bottom trace shows the activation probability of AMBER computed using the Boltzmann function fitted to the patch clamp data (see Fig. 2B). Shaded region is the corresponds to the best fit monoexponential weighted by the same activation curve. The dark blue pulses are bioluminescent signals (median, n=8) produced by AMBER and closely follows the 450ms decay curve (dark blue dotted line). The light blue dotted line is the low frequency envelope of the bioluminescent signal and indicates the rate of substrate consumption and production.(B) The on and off time constant of AMBER as determined from the electrical patch clamp current recordings. Depolarizing current transients were obtained by applying 20ms voltage steps from −60 mV to 60 mV at an incremental step of 20 mV from the holding potential of −60 mV. The fast component of luminescence change is expected to correspond to the charge movement and therefore the time constant [64]. (C) A proposed model for the activation of AMBER upon membrane depolarization. At the resting state, the transfer of FMNH2 from FRP to eluxAB is prevented thereby shutting off the biochemical path of the light reaction. VSD S4 transmembrane domain undergoes a conformational change upon membrane depolarization causing FMNH2 transfer from FRP to eluxAB domain.
2.8. Molecular Modeling Studies
The most probable cause for the enzymatic switching can be attributed to the position of YPet relative to eluxAB because switching their position produced a dim signal, First, we established a molecular basis for the eluxAB function based on the structural details underpinning the function of V. harveyi luxAB (vh-luxAB): FMN complex [65]. vh-luxAB bears large sequence similarity to eLuxAB (Figure S9). Predictive models computed using the I-TASSER [66, 67, 68] showed how eluxAB folding affects the FMN binding pocket geometry. In particular, we find that association constraint between the α and β subunits is critical for the enzyme function as this directly affects the FMN pocket geometry (See the SI Section 1.7 and Figure S14). Hence, the interface distance constraints of vh-luxAB subunits in the FMN unbound/bound states were applied to simulate the association between α and β domains of eluxAB in unbound/bound states (Table S3). Both interface distance constraints and the sequence alignment information were used to accurately predict the structure of eluxAB, eluxAB-YPet, and YPeteluxAB (see Figure S15) in the FMN unbound/bound states. We found that the FMN binding pocket of YPet-eluxAB in the unbound state was severely contracted. The solvent accessible surface area (SASA) was ≈34.5% less than that of the bound state as opposed to the ≈9.7% contraction for that of the eluxABYPet. These results are in-line with the experimentally observed bright bioluminescence for FV-E-Y and the dim bioluminescence for FV-Y-E and V-Y-E constructs. Furthermore, there was little change in the Förster distance (mean distance between FMN pocket residues and the YPet chromophore residues) for both BRET pair models, which explains the minor contribution of BRET in the differential signals. In contrast, we did not see any appreciable change in the FMN pocket geometry for the eluxAB (SASA≈1%) and this explains for the negligible change in the bioluminescent signal of V-E construct upon depolarization. Mean solvation free energies (See the SI Section 1.7 and Figure S16) of all the predicted structures were estimated using the approach described elsewhere[69].
Similar to the vh-luxAB: FMN complex [65], the SASA of FMN pocket in the unbound state was predicted to be smaller than that of the bound state for both BRET pair models. Hence, thermal mobility of the FMN pocket residues has to play a critical part in the eluxAB: FMN binding. Therefore, we compared the predicted values of normalized B-factor, an indicator of thermal mobility, for key residues of the FMN binding pocket in the unbound state (see Figure S15b). The eLuxAB structure exhibits high thermal mobility for ARG107, LEU109, TYR110, GLU175 and SER176. Mutation of GLU175 was reported to reduce activity of vh-luxAB to < 1% [70] and plays an important role in light reaction kinetics. For most residues, the thermal mobility is markedly different from the eLuxAB structure for YPeteLuxAB and to a lesser extent in eLuxAB-YPet. Taken together these results shed light on why the enzymatic activity is affected when the position of eLuxAB and YPet are switched. Based on these observations we propose the mechanism shown in Figure 3C for the enzymatic switching of AMBER.
2.9. In vivo Voltage Activity Recorded Using AMBER
We created C. elegans transgenic lines to test AMBER function in vivo. We chose C. elegans as the model organism because of the optically transparent properties of its tissues, and the slow kinetics of the muscle and neuronal voltage signals (about a few seconds). We subcloned AMBER cDNAs in C. elegans expression vectors (See the SI Section 1.2). Myosin-2 heavy chain gene, myo-2[71] and the α-tubulin gene, mec-7 promoters [72, 73] were used to drive the transgene expression in pharyngeal muscles and mechanosensory touch neurons, respectively (See the SI Section 1.8). We genetically targeted specific tissues of interest and recorded bioluminescent voltage signals using a custom-built imaging setup that allows tracking the animal positions simultaneously (See the SI Section 1.9).
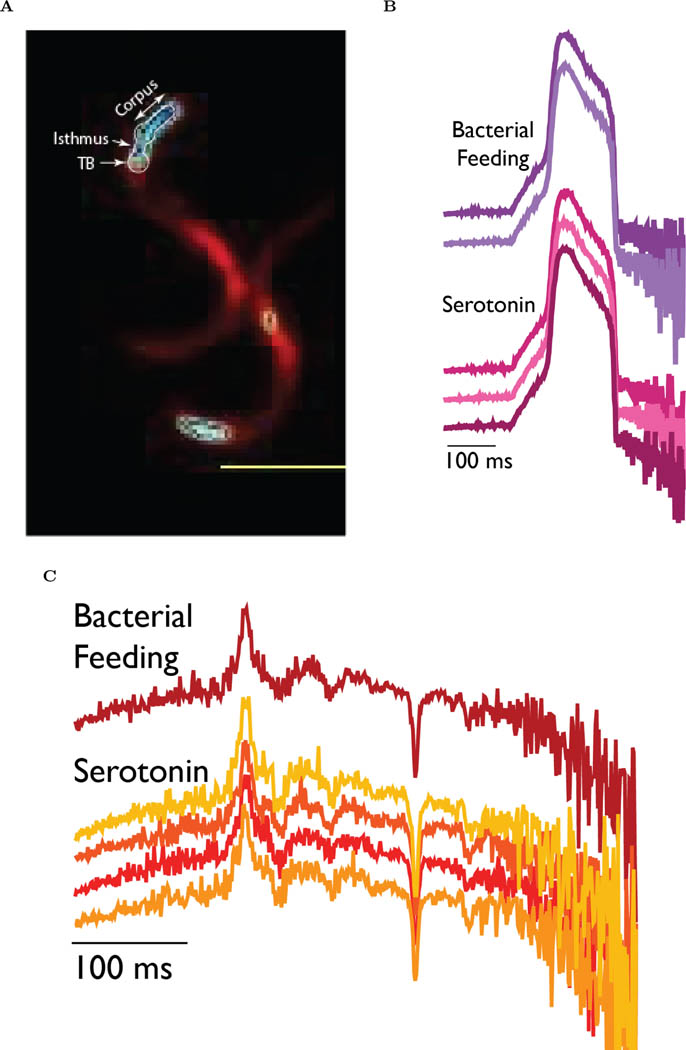
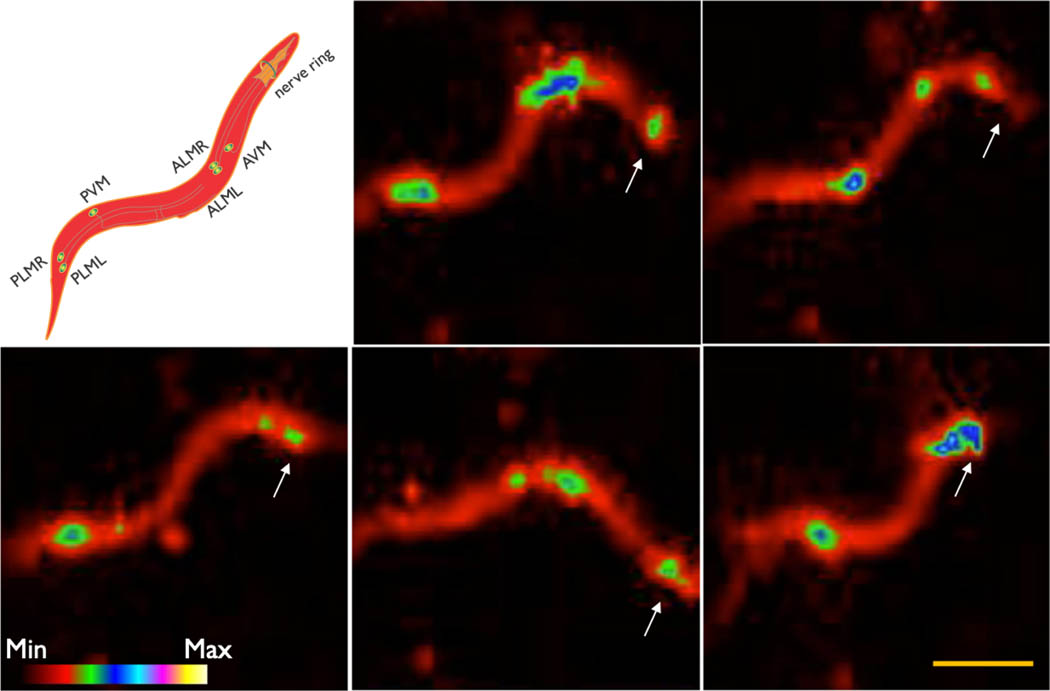
First, we wanted to conform if AMBER is capable of reporting physiological voltage signals. C. elegans uses pharyngeal pumping to concentrate its food and that starvation stimulates pumping action. Electrophysiological response of a C. elegans pharynx was generally characterized using extracellular (Electropharyngeogram, ‘EPG’) and intracellular (Terminal bulb action potential, ‘AP’) electrical recordings under starving conditions, which stimulates pumping[74]. Recently, the fast transients of EPG and AP traces were recorded using a fluorescent voltage sensor [75]. We recorded voltage activity of the entire pharynx and the terminal bulb under serotonin-induced simulated starvation and bacterial feeding conditions (Figure 4A and the SI Movie SM1). Serotonin increases the rate of feeding in C. elegans [76] and is generally considered to provide neuromuscular food signals to the animals [77, 78, 79]. Voltage imaging was done for tens of seconds integrating the signals for 500 msec per frame (See the SI Section 1.10). This image capture speed is too slow to record the fast muscle kinetics of C. elegans pharynx (≈300 msec for EPG and ≈225 msec for terminal bulb AP). We, therefore, modified a recently developed technique [80] using sparse sampling approach to reconstruct the terminal bulb AP (Figure 4B) and the EPG (Figure 4C) of different animals under starvation and bacterial feeding conditions. Our approach relies on a priori knowledge of the signal shape for EPG[81] and AP[82] obtained electrically at high temporal resolution (See the SI Section 1.10 for the ground truth signals of EPG and the terminal bulb AP). We reconstructed the EPG trace with sufficient accuracy so the dominant positive (TB depolarization) and negative (Corpus repolarization) transients are clearly resolved. However, some small positive and negative transients were not resolved fully indicating the insufficiency of the representative samples for reconstruction. On the other hand, SNR of the reconstructed AP traces was good enough to resolve the depolarization and repolarization kinetics. Based on these findings, we confirm the ability of AMBER to sense physiological voltage signals during pharyngeal pumping events in C. elegans. Next, we expressed AMBER at sufficient levels in the mechanosensory touch neurons (ALMR, ALML, AVM, PLMR, PLML, PVM) and the anterior nerve ring of C. elegans (See the SI Section 1.11). Signals from the touch neurons did not show any activity when the animals were moving unilaterally in the forward direction but showed bursts of activity while making spatially restricted movements and frequent reversals (Figure 5 and the SI movies SM2 and SM3) or during collisions (Figure S7). Earlier studies reported that mechanosensory touch neurons respond to changes in force, but not to the exertion of a constant force [83, 84]. We observed varying levels of activities of the touch neurons during collisions (Figure S7) that can exert transient differential forces due to momentum transfer. The underlying mechanism of differential force exertion during frequent reversals and restricted movements is not fully understood. Nevertheless, these motion trajectories on the hydrated agar bed were earlier reported to be involved in detecting food [85]. A huge advantage of AMBER is the ability to simultaneously record the activity of a neural circuit from multiple worms moving in different directions within the field of view, which is not possible using fluorescence (Figure S8 and the SI Movie SM4).
Figure 4:
In vivo voltage imaging of the pharyngeal muscle activity of adult C. elegans under stimulated starvation and bacterial feeding conditions. (A) Bioluminescent voltage signals recorded from the pharynx of transgenic animals expressing AMBER. The scale bar length is 500μm. (B) Terminal bulb action potential traces reconstructed using experimental recordings (N= 200 samples) obtained from the individual animals under different conditions. A modified sparse sampling approach was applied to reconstruct the traces. Bootstrap sampling (N= 3000 samples) of the recorded data increased the signal-to-noise ratio of the representation of the ground truth signal. (C) Electropharyngeogram kinetics reconstructed using experimental recordings (N= 200 samples) obtained from the individual animals under different conditions. A modified sparse sampling approach was applied to reconstruct the traces after Bootstrap sampling (N= 3000 samples).The different colors of the traces correspond to different animals.(n= 5 animals for Serotonin treatment; n=3 animals for bacterial feeding; Bootstrap Sampling)
Figure 5:
Snapshots of the touch neurons activities of a C. elegans worm recorded using AMBER. The worm takes different shapes as shown while performing frequent reversal movements causing graded activity in the mechanosensory touch-neurons (PLM, PVM, ALM, AVM and the anterior nerve ring neurons). Arrowheads indicate the anterior of the worm (n=3 animals). The scale bar length is 500μm.
3. Discussion
We report the creation of a first generation voltage-sensitive bioluminescent probe (AMBER), which uses a Ciona VSD, a synthetic enhanced bacterial luciferase, eluxAB, and a β barrel domain of the fluorescent YPet. The system is autonomous and enables the expression of the probe and its substrate genetically, circumventing a major drawback of other bioluminescent systems. We engineered the placement of various domains to achieve a high SNR and efficient switching of the enzymatic activity. Since AMBER switches its enzymatic activity reversibly with membrane potential, we performed several experiments to understand the switching mechanism. We shed light on the mechanism of enzymatic modulation using the data obtained from the computational models, cellular imaging, and spectroscopy measurements. Three factors frame the discussion around the mechanism behind enzymatic switching: the bioluminesence reaction mechanism, the role of YPet in the mechanism, and the role of the substrates. The bioluminescence reaction pathway proceeds by the FMNH2 forming a complex with the luciferase enzyme. The Enzyme-Flavin complex then reacts with molecular oxygen to produce a hyrdoxyperoxyflavin intermediate, which the subsequently reacts with the aldehyde to eventually emit a photon[86, 87]. The order of the reaction points to the relative importance of FMNH2 in the switching mechanism. The role of YPet is best understood by considering the results from the experiments performed on the dark mutant (FV-E-Dark) (see figure 2A). Results from the dark mutant (FV-E-Dark) conclusively proved the only necessary factor in the switching mechanism is the physical structure of YPet and not the resonance energy transfer. Furthermore, molecular modeling studies showed the position of YPet relative to eluxAB modulates the FMNH2 binding pocket. A detailed analysis of the molecular model supporting this argument from energetic considerations is provided in the SI. The only other method of modulating the enzymatic activity would be to modulate the access of the aldehyde by the voltage. However, as shown in figure 1C, the construct VY-E with and without the substrate producing proteins (FRP and luxCDE) showed similar activity. More importantly, fusing FRP to the N-terminus of this construct greatly reduced its activity (see FV-Y-E in figure 1C). This reduction in activity suggests the increased availability of FMN does not improve the light output, which would have been the case if aldehyde was the reason for the switching mechanism. he VY-E (or non-optimal) constructs serve as the ideal platform to compare the effect of aldehyde since they exhibit reduced performance with respect to FMNH2 (as evidenced by the poor performance of the FV-Y-E construct) and additionally, eluxAB has better access to the aldehyde since it is more exposed to the cytosol. Based on the experimental data and molecular modeling we propose the following as a plausible mechanism for the voltage switching behavior of AMBER. In the resting state, FMN binding to eluxAB is inhibited due to contraction of the FMN pocket accompanied by the native folding of the eluxAB-YPet. Upon membrane depolarization, the movement of the S4 transmembrane helix of the VSD reorients the YPet that coordinates with the eluxA movement through association constraints. These coordinated movements of eluxAB-YPet BRET pair perturb the initially contracted FMN pocket thereby opening it to accommodate an incoming FMNH2. A schematic representation of this molecular mechanism is shown in Figure 3C.
We performed all optical electrophysiology to determine the switching kinetics of AMBER. The slow inactivation and depolarized state of the cell even in resting conditions made it difficult to directly measure the time constants. However, determining the time constants of the gating currents enabled us to estimate the fast component of the switching kinetics, since the charge kinetics closely reflects the fast component[64]. By analysing the slow envelope modulation resulting from substrate recycling we were able to set a upper bound on the slow component to be 200–300 ms. By using a sparse sampling approach, we were able to effectively reconstruct the voltage signals of much faster events with a sample size of a couple of hundred recordings in vivo (See Figures 4B & 4C). This enabled detecting faster kinetics even though the probe might be intrinsically slower.
We have shown for the first time the ability to modulate the enzymatic activity of a voltage-gated luciferase by varying the membrane potential. We propose that the voltage imaging approach described here will have broad applicability, especially when coupled with signal reconstruction techniques. One key advantage of a bioluminescent voltage reporter is the ability to monitor voltage activity for a prolonged (days) period. Current fluorescent reporters of voltage activity usually require high photon flux due to their limited sensitivity resulting in a rather short experimental duration (15 minutes- 1 hour)[2]. While the kinetics of AMBER needs improvement for single neuron experiments, it highlights the advantage of bioluminescence for long-term voltage imaging. Additionally, tools like AMBER will help decipher the critical role of the membrane potential in many biological processes ranging from cancer biology to wound healing [29], which are intrinsically much slower (few hundred milliseconds to seconds). A particularly important area of application is the study of cardiomyocytes derived from stem cells as a potent in vitro drug screening platform[88, 89, 47]. Additionally, monitoring changes in membrane potential over a long time period is essential to studying the role of membrane potential in regulating downstream gene expression by cells interacting via gap junctions [90], which in turn has major implications in cell proliferation and function.
4. Conclusions
We engineered an autonomous voltage-sensitive bioluminescent reporter based on the bacterial luciferase system. The ability to genetically encode the luciferase and the luciferin overcomes many challenges inherent with firefly and the deep-sea shrimp bioluminescent systems. Moreover, by extensive molecular engineering of the probe, we obtained bright signals upon depolarization. AMBER exhibits a voltage-dependent enzymatic switching not observed in other similar systems (e.g. LOTUS-V). We successfully expressed and demonstrated AMBER function both in HEK293 cells and in C. elegans. We report the activities of the mechanosensory neural circuit and the pharyngeal muscle pumping from multiple animals using AMBER. We believe this will greatly enhance the ability to track the behavior of multiple animals, while simultaneously monitoring the voltage activity. Tools like AMBER can help unravel the critical role played by the membrane potential in cell proliferation, differentiation and apoptosis.
Supplementary Material
Acknowledgements
This work was partially funded by the Otis Williams Fellowship (PS), the NSF Neuronex (Award 1934288 LT), the National Eye Institute (EY008117 and EY010852; CM), the National Institute of Health (1R01HD-082347 and 1R01HD081266; JR), the American Heart Foundation (19POST34430179; DT) and the Institute of Collaborative Biotechnologies at the UCSB. The authors are grateful to Sumita Pennathur, UCSB for generously lending the EMCCD camera and Spencer Smith, UCSB for helping build the patch clamp set up. The authors thank Dan Close and Gary Sayler at 490 Biotech for providing us the pCMVlux plasmid. We also acknowledge Ute Hochgeschwender (Central Michigan University) and Nathan Shaner (UC San Diego) for insightful discussions.
Footnotes
Supporting Information
Experimental procedures, matlab scripts and characterization data can be found in the supplementary information available from the Wiley Online Library or from the author.
Contributor Information
Dr. Prasanna Srinivasan, Department of Electrical and Computer Engineering, University of California Santa Barbara, CA 93106; Center for Bioengineering, Institute for Collaborative Biotechnologies, University of California Santa Barbara, CA 93106.
Nicole M Griffin, Department of Electrical and Computer Engineering, University of California, Santa Barbara, CA, 93106, USA.; Center for Bioengineering, Institute for Collaborative Biotechnologies, University of California, Santa Barbara, CA, 93106, USA.
Dr. Dhananjay Thakur, Department of Molecular, Cellular and Developmental Biology, University of California Santa Barbara, CA 93106 The Neuroscience Research Institute, University of California Santa Barbara, CA 93106.
Dr. Pradeep Joshi, Department of Molecular, Cellular and Developmental Biology, University of California Santa Barbara, CA 93106
Alex Nguyen-Le, Department of Electrical and Computer Engineering, University of California Santa Barbara, CA 93106; Current address: Department of Electrical Engineering, University of Pennsylvania, Philadelphia, PA.
Sean McCotter, Department of Electrical and Computer Engineering, University of California Santa Barbara, CA 93106.
Akshar Jain, Department of Electrical and Computer Engineering, University of California Santa Barbara, CA 93106.
Mitra Saeidi, Department of Electrical and Computer Engineering, University of California Santa Barbara, CA 93106.
Dr. Prajakta Kulkarni, Department of Electrical and Computer Engineering, University of California Santa Barbara, CA 93106
Jaclyn T. Eisdorfer, College of Creative Studies,University of California Santa Barbara, CA 93106 Current address: Dept. of Bioengineering, Temple University, Philadelphia, PA 19122
Prof. Dr. Joel Rothman, Department of Molecular, Cellular and Developmental Biology, University of California Santa Barbara, CA 93106
Prof. Dr. Craig Montell, Department of Molecular, Cellular and Developmental Biology, University of California Santa Barbara, CA 93106 The Neuroscience Research Institute, University of California Santa Barbara, CA 93106.
Prof. Dr. Luke Theogarajan, Department of Electrical and Computer Engineering, University of California Santa Barbara, CA 93106; Center for Bioengineering, Institute for Collaborative Biotechnologies, University of California Santa Barbara, CA 93106.
References
- [1].Mehta S, Zhang J, Annual Review of Biochemistry 2011, 80 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lin MZ, Schnitzer MJ, Nature Neuroscience 2016, 19, 9 1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Xu Y, Zou P, Cohen AE, Current Opinion in Chemical Biology 2017, 39 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Qian Y, Piatkevich KD, Mc Larney B, Abdelfattah AS, Mehta S, Murdock MH, Gottschalk S, Molina RS, Zhang W, Chen Y, Wu J, Drobizhev M, Hughes TE, Zhang J, Schreiter ER, Shoham S, Razansky D, Boyden ES, Campbell RE, Nature Methods 2019, 16, 2 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Tian L, Andrew Hires S, Looger LL, Imaging neuronal activity with genetically encoded calcium indicators, volume 7, Cold Spring Harb protoc, 2012. [DOI] [PubMed] [Google Scholar]
- [6].Bischof H, Rehberg M, Stryeck S, Artinger K, Eroglu E, Waldeck-Weiermair M, Gottschalk B, Rost R, Deak AT, Niedrist T, Vujic N, Lindermuth H, Prassl R, Pelzmann B, Groschner K, Kratky D, Eller K, Rosenkranz AR, Madl T, Plesnila N, Graier WF, Malli R, Nature Communications 2017, 8, 1 1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Shen Y, Wu SY, Rancic V, Aggarwal A, Qian Y, Miyashita SI, Ballanyi K, Campbell RE, Dong M, Communications Biology 2019, 2, 1 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Liang R, Broussard GJ, Tian L, ACS Chemical Neuroscience 2015, 6, 1 84. [DOI] [PubMed] [Google Scholar]
- [9].Okumoto S, Looger LL, Micheva KD, Reimer RJ, Smith SJ, Frommer WB, Proceedings of the National Academy of Sciences of the United States of America 2005, 102, 24 8740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Marvin JS, Borghuis BG, Tian L, Cichon J, Harnett MT, Akerboom J, Gordus A, Renninger SL, Chen TW, Bargmann CI, Orger MB, Schreiter ER, Demb JB, Gan WB, Hires SA, Looger LL, Nature Methods 2013, 10, 2 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yang HH, St-Pierre F, Journal of Neuroscience 2016, 36, 39 9977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Abdelfattah AS, Farhi SL, Zhao Y, Brinks D, Zou P, Ruangkittisakul A, Platisa J, Pieribone VA, Ballanyi K, Cohen AE, Campbell RE, Journal of Neuroscience 2016, 36, 8 2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kannan M, Vasan G, Huang C, Haziza S, Li JZ, Inan H, Schnitzer MJ, Pieribone VA, Nature Methods 2018, 15, 12 1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yang W, Yuste R, Nature Methods 2017, 14, 4 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Stirman JN, Smith IT, Kudenov MW, Smith SL, Nature Biotechnology 2016, 34, 8 857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wäldchen S, Lehmann J, Klein T, van de Linde S, Sauer M, Scientific Reports 2015, 5, 1 15348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Benninger RKP, Piston DW, Current Protocols in Cell Biology 2013, 59, 1 4.11.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Fosque BF, Sun Y, Dana H, Yang CT, Ohyama T, Tadross MR, Patel R, Zlatic M, Kim DS, Ahrens MB, Jayaraman V, Looger LL, Schreiter ER, Science 2015, 347, 6223 755. [DOI] [PubMed] [Google Scholar]
- [19].Zhao Y, Araki S, Wu J, Teramoto T, Chang YF, Nakano M, Abdelfattah AS, Fujiwara M, Ishihara T, Nagai T, Campbell RE, Science 2011, 333, 6051 1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yang HH, St-Pierre F, Sun X, Ding X, Lin MZ, Clandinin TR, Cell 2016, 166, 1 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wei Z, Lin BJ, Chen TW, Daie K, Svoboda K, Druckmann S, A comparison of neuronal population dynamics measured with calcium imaging and electrophysiology, 2019, URL 10.1101/840686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Burgoyne RD, Nature Reviews Neuroscience 2007, 8, 3 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Grienberger C, Konnerth A, Neuron 2012, 73, 5 862. [DOI] [PubMed] [Google Scholar]
- [24].Knöpfel T, Nature Reviews Neuroscience 2012, 13, 10 687. [DOI] [PubMed] [Google Scholar]
- [25].Antic SD, Empson RM, Knöpfel T, Journal of Neurophysiology 2016, 116, 1 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Piatkevich KD, Bensussen S, Tseng H.-a., Shroff SN, Physiology & behavior 2017, 176, 10 139.28363838 [Google Scholar]
- [27].Abdelfattah AS, Kawashima T, Singh A, Novak O, Liu H, Shuai Y, Huang YC, Grimm JB, Patel R, Friedrich J, Mensh BD, Paninski L, Macklin JJ, Podgorski K, Lin BJ, Chen TW, Turner GC, Liu Z, Koyama M, Svoboda K, Ahrens MB, Lavis LD, Schreiter ER, bioRxiv 2018, 365 699. [Google Scholar]
- [28].Cohen AE, Venkatachalam V, Annual Review of Biophysics 2014, 43, 1 211. [DOI] [PubMed] [Google Scholar]
- [29].Kadir LA, Stacey M, Barrett-Jolley R, Frontiers in Physiology 2018, 9, NOV 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bando Y, Sakamoto M, Kim S, Ayzenshtat I, Yuste R, Cell Reports 2019, 26, 3 802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lorenz WW, McCann RO, Longiaru M, Cormier MJ, Proceedings of the National Academy of Sciences of the United States of America 1991, 88, 10 4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Markova SV, Golz S, Frank LA, Kalthof B, Vysotski ES, Journal of Biological Chemistry 2004, 279, 5 3212. [DOI] [PubMed] [Google Scholar]
- [33].Thompson EM, Nagata S, Tsuji FI, Proceedings of the National Academy of Sciences of the United States of America 1989, 86, 17 6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Verhaegent M, Christopoulos TK, Analytical Chemistry 2002, 74, 17 4378. [DOI] [PubMed] [Google Scholar]
- [35].Wood KV, Lam YA, McElroy WD, Journal of bioluminescence and chemiluminescence 1989, 4, 1 289. [DOI] [PubMed] [Google Scholar]
- [36].Kaskova ZM, Dörr FA, Petushkov VN, Purtov KV, Tsarkova AS, Rodionova NS, Mineev KS, Guglya EB, Kotlobay A, Baleeva NS, Baranov MS, Arseniev AS, Gitelson JI, Lukyanov S, Suzuki Y, Kanie S, Pinto E, Mascio PD, Waldenmaier HE, Pereira TA, Carvalho RP, Oliveira AG, Oba Y, Bastos EL, Stevani CV, Yampolsky IV, Science Advances 2017, 3, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Brodl E, Winkler A, Macheroux P, Computational and Structural Biotechnology Journal 2018, 16 551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kotlobay AA, Sarkisyan KS, Mokrushina YA, Marcet-Houben M, Serebrovskaya EO, Markina NM, Gonzalez Somermeyer L, Gorokhovatsky AY, Vvedensky A, Purtov KV, Petushkov VN, Rodionova NS, Chepurnyh TV, Fakhranurova LI, Guglya EB, Ziganshin R, Tsarkova AS, Kaskova ZM, Shender V, Abakumov M, Abakumova TO, Povolotskaya IS, Eroshkin FM, Zaraisky AG, Mishin AS, Dolgov SV, Mitiouchkina TY, Kopantzev EP, Waldenmaier HE, Oliveira AG, Oba Y, Barsova E, Bogdanova EA, Gabaldón T, Stevani CV, Lukyanov S, Smirnov IV, Gitelson JI, Kondrashov FA, Yampolsky IV, Proceedings of the National Academy of Sciences 2018, 115, 50 12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Shieh YW, Minguez P, Bork P, Auburger JJ, Guilbride DL, Kramer G, Bukau B, Science 2015, 350, 6261 678. [DOI] [PubMed] [Google Scholar]
- [40].Gregor C, Gwosch KC, Sahl SJ, Hell SW, Proceedings of the National Academy of Sciences of the United States of America 2018, 115, 5 962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Close DM, Patterson SS, Ripp S, Baek SJ, Sanseverino J, Sayler GS, PLoS ONE 2010, 5, 8 12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hollis RP, Lagido C, Pettitt J, Porter AJ, Killham K, Paton GI, Glover LA, FEBS Letters 2001, 506, 2 140. [DOI] [PubMed] [Google Scholar]
- [43].Gregor C, Pape JK, Gwosch KC, Gilat T, Sahl SJ, Hell SW, Proceedings of the National Academy of Sciences 2019, 116, 52 26491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Li Q, Wanderling S, Paduch M, Medovoy D, Singharoy A, Mcgreevy R, Villalba-Galea CA, Hulse RE, Roux B, Schulten K, Kossiakoff A, Perozo E, Nature Structural and Molecular Biology 2014, 21, 3 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Nguyen AW, Daugherty PS, Nature Biotechnology 2005, 23, 3 355. [DOI] [PubMed] [Google Scholar]
- [46].Dimitrov D, He Y, Mutoh H, Baker BJ, Cohen L, Akermann W, Knöpfel T, PLoS ONE 2007, 2, 5 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Inagaki S, Tsutsui H, Suzuki K, Agetsuma M, Arai Y, Jinno Y, Bai G, Daniels MJ, Okamura Y, Matsuda T, Nagai T, Scientific Reports 2017, 7, 1 42398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Inagaki S, Agetsuma M, Ohara S, Iijima T, Yokota H, Wazawa T, Arai Y, Nagai T, Scientific Reports 2019, 9, 1 7460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, MacHleidt T, Robers MB, Benink HA, Eggers CT, Slater MR, Meisenheimer PL, Klaubert DH, Fan F, Encell LP, Wood KV, ACS Chemical Biology 2012, 7, 11 1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Cui B, Zhang L, Song Y, Wei J, Li C, Wang T, Wang Y, Zhao T, Shen X, PLoS ONE 2014, 9, 10 107885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Machleidt T, Woodroofe CC, Schwinn MK, Méndez J, Robers MB, Zimmerman K, Otto P, Daniels DL, Kirkland TA, Wood KV, ACS Chemical Biology 2015, 10, 8 1797. [DOI] [PubMed] [Google Scholar]
- [52].Lei B, Tu SC, Biochemistry 1998, 37, 41 14623. [DOI] [PubMed] [Google Scholar]
- [53].Tu SC, Photochemical and Photobiological Sciences 2008, 7, 2 183. [DOI] [PubMed] [Google Scholar]
- [54].Jawanda N, Ahmed K, Tu SC, Biochemistry 2008, 47, 1 368. [DOI] [PubMed] [Google Scholar]
- [55].Lambert TJ, Nature Methods 2019, 16 227. [DOI] [PubMed] [Google Scholar]
- [56].Wielgus-Kutrowska B, Narczyk M, Buszko A, Bzowska A, Clark PL, Journal of Physics Condensed Matter 2007, 19, 28 285223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Schaefer PM, Kalinina S, Rueck A, von Arnim CA, von Einem B, Cytometry Part A 2019, 95, 1 34. [DOI] [PubMed] [Google Scholar]
- [58].Sung U, Sepehri-Rad M, Piao HH, Jin L, Hughes T, Cohen LB, Baker BJ, PLoS ONE 2015, 10, 11 141585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Akemann W, Lundby A, Mutoh H, Knöpfel T, Biophysical Journal 2009, 96, 10 3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Jin L, Han Z, Platisa J, Wooltorton JR, Cohen LB, Pieribone VA, Neuron 2012, 75, 5 779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Caterina MJ, Schumacher MA, Tominaga M, Rosen TA, Levine JD, Julius D, Nature 1997, 389, 6653 816. [DOI] [PubMed] [Google Scholar]
- [62].Cao E, Cordero-Morales JF, Liu B, Qin F, Julius D, Neuron 2013, 77, 4 667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Lin JY, Knutsen PM, Muller A, Kleinfeld D, Tsien RY, Nature Neuroscience 2013, 16, 10 1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Villalba-Galea CA, Sandtner W, Dimitrov D, Mutoh H, Knöpfel T, Bezanilla F, Biophysical journal 2009, 96, 2 L19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Campbell ZT, Weichsel A, Montfort WR, Baldwin TO, Biochemistry 2009, 48, 26 6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y, Nature Methods 2014, 12, 1 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Roy A, Kucukural A, Zhang Y, Nature Protocols 2010, 5, 4 725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Zhang Y, BMC Bioinformatics 2008, 9 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Eisenberg D, Mclachlan AD, Nature 1986, 319, 6050 199. [DOI] [PubMed] [Google Scholar]
- [70].Hosseinkhani S, Szittner R, Meighen EA, Biochemical Journal 2005, 385, 2 575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Okkema PG, Harrison SW, Plunger V, Aryana A, Fire A, Genetics 1993, 135, 2 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Hamelin M, Scott IM, Way JC, Culotti JG, EMBO Journal 1992, 11, 8 2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Zhang Y, Ma C, Delohery T, Nasipak B, Foat BC, Bounoutas A, Bussemaker HJ, Kim SK, Chalfle M, Nature 2002, 418, 6895 331. [DOI] [PubMed] [Google Scholar]
- [74].Cook A, Franks CJ, Holden-Dye L, WormBook : the online review of C. elegans biology 2006, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Hashemi NA, Bergs AC, Schüler C, Scheiwe AR, Costa WS, Bach M, Liewald JF, Gottschalk A, Proceedings of the National Academy of Sciences of the United States of America 2019, 116, 34 17051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Avery L, Horvitz HR, Journal of Experimental Zoology 1990, 253, 3 263. [DOI] [PubMed] [Google Scholar]
- [77].Horvitz HR, Chalfie M, Trent C, Sulston JE, Evans PD, Science 1982, 216, 4549 1012. [DOI] [PubMed] [Google Scholar]
- [78].Raizen DM, Lee RY, Avery L, Genetics 1995, 141, 4 1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Niacaris T, Avery L, Journal of Experimental Biology 2003, 206, 2 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Mano O, Creamer MS, Matulis CA, Salazar-Gatzimas E, Chen J, Zavatone-Veth JA, Clark DA, Nature Communications 2019, 10, 1 4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Avery L, Raizen D, Lockery S, Methods in Cell Biology 1995, 48, C 251. [PMC free article] [PubMed] [Google Scholar]
- [82].Davis MW, Fleischhauer R, Dent JA, Joho RH, Avery L, Science 1999, 286, 5449 2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Goodman MB, Science 2004, 306, 5695 427. [DOI] [PubMed] [Google Scholar]
- [84].Krieg M, Dunn AR, Goodman MB, BioEssays 2015, 37, 3 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Goodman MB, WormBook : the online review of C. elegans biology 2006, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Francisco WA, Abu-Soud HM, DelMonte AJ, Singleton DA, Baldwin TO, Raushel FM, Biochemistry 1998, 37, 8 2596. [DOI] [PubMed] [Google Scholar]
- [87].Abu-Soud H, Mullins LS, Baldwin TO, Raushel FM, Biochemistry 1992, 31, 15 3807. [DOI] [PubMed] [Google Scholar]
- [88].Leyton-Mange JS, Mills RW, Macri VS, Jang MY, Butte FN, Ellinor PT, Milan DJ, Stem cell reports 2014, 2, 2 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Shinnawi R, Huber I, Maizels L, Shaheen N, Gepstein A, Arbel G, Tijsen AJ, Gepstein L, Stem cell reports 2015, 5, 4 582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Levin M, Molecular Biology of the Cell 2014, 25, 24 3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.