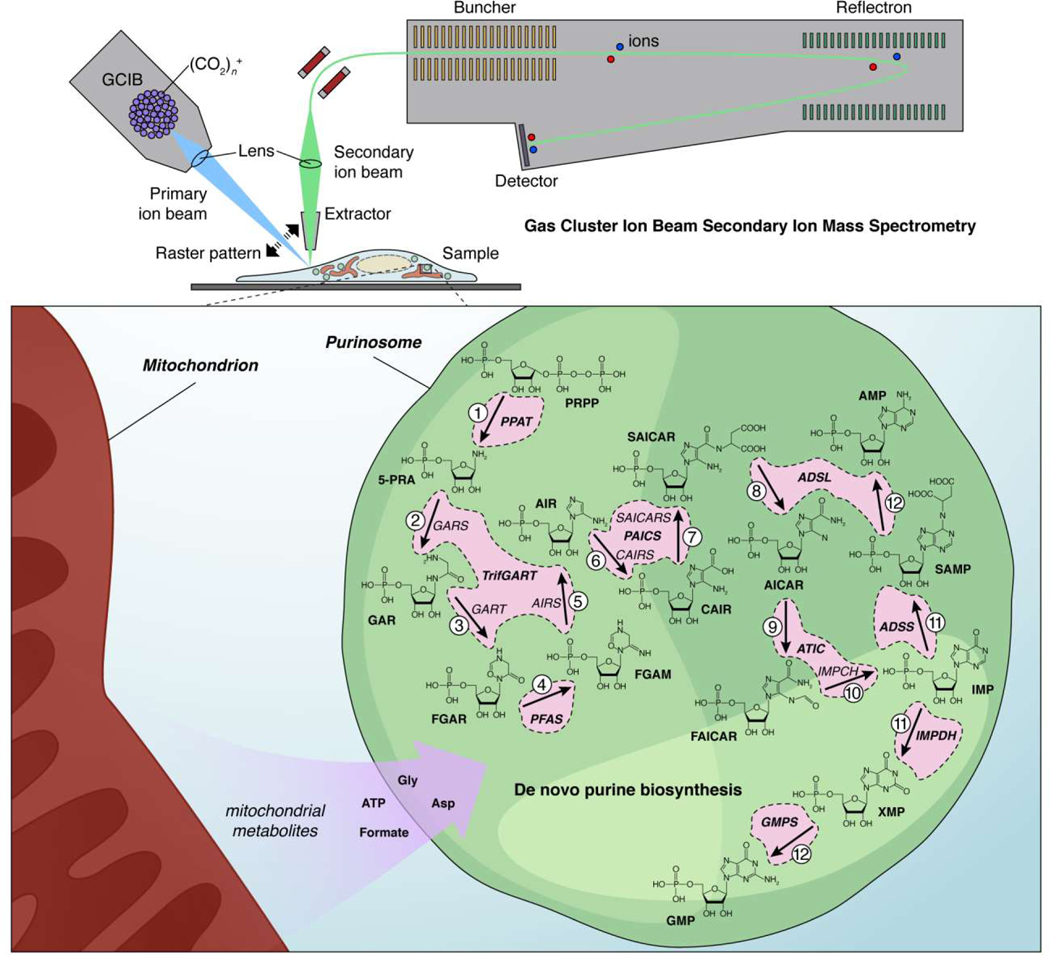
Figure 3. Mass spectrometry imaging reveals substrate channeling by the purinosome, a liquid-like multi-enzyme assembly.
(Upper panel) In gas cluster ion beam secondary ion mass spectrometry (GCIB-SIMS), clusters containing many thousands of ionized gas molecules (e.g., [CO2]n+, where n>10,000) are a fired at high velocity (primary ion beam) to liberate molecular ions from the surface of a frozen sample (e.g., a cell). These molecular ions are then extracted and focused into a secondary ion beam that travels into the detector. The primary ion beam is focused into a small spot (e.g., 1 μm × 1 μm × 400 nm) and raster scanned at different depths across the sample to create a 3D “image” where each voxel corresponds to a mass spectrum. (Lower panel) Pareek et al. used this technology to directly visualize the substrate-channeling behavior of the purinosome, a multi-enzyme condensate that controls de novo purine biosynthesis, in HeLa cells [68]. Based on their studies, the authors were ultimately able to conclude that active purinosomes comprise all 9 enzymes responsible for catalyzing the 14-step synthesis of AMP and GMP from PRPP in a highly channeled process, insulated from the bulk cytosol, and that localization near mitochondria further allows purinosomes to effectively channel necessary cofactors, produced via mitochondrial metabolism, directly into the de novo purine biosynthesis pathway.