Figure 11.
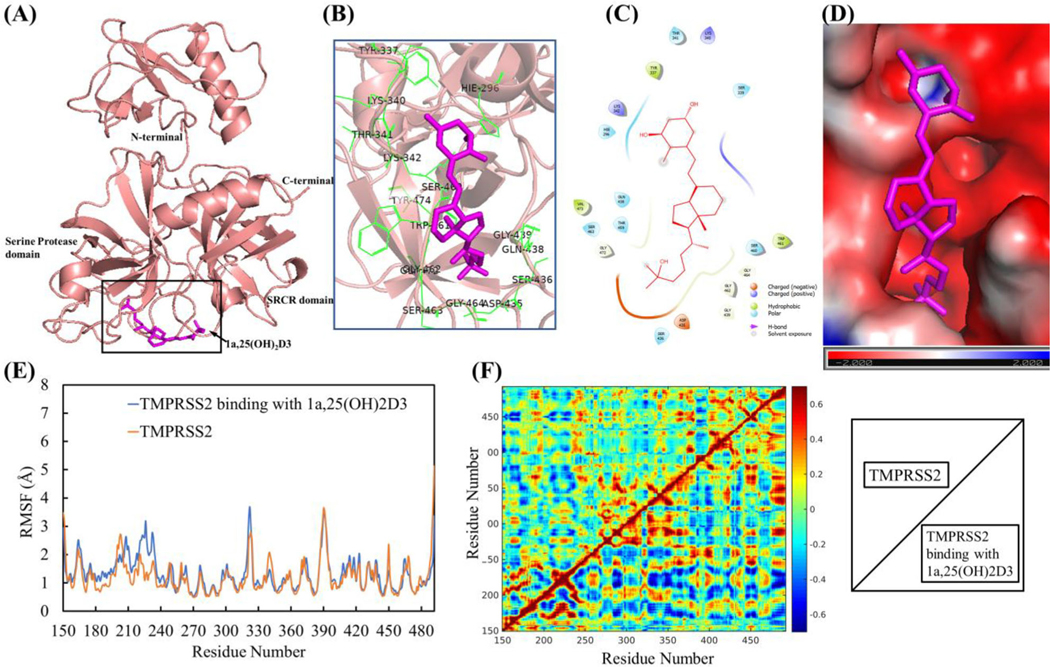
1a,25(OH)2D3 (1,25(OH)2D3) interactions with TMPRSS2 resulted in the conformational and dynamical motion changes. (A) The representative TMPRSS2 binding mode with 1,25(OH)2D3 from equilibrated MD simulation (image made with PyMOL (v2.4.0, https://pymol.org/2/)). (B) 3D representation of 1,25(OH)2D3 interaction with binding region of TMPRSS2 (image made with PyMOL (v2.4.0, https://pymol.org/2/)). (C) 2D representation of 1,25(OH)2D3 interaction with binding region of TMPRSS2 (image made with Maestro (v12.4, https://www.schrodinger.com/products/maestro)). (D) Electrostatic potential of TMPRSS2 at its binding region with 1,25(OH)2D3 (in magenta). Red: negative electrostatic potential; Blue: positive electrostatic potential and White: neutral electrostatic potential. (image made with PyMOL (v2.4.0,https://pymol.org/2/)). (E) RMSF of TMPRSS2 with and without 1,25(OH)2D3 binding (Duplicate and data shown in average). (F) DCCM dynamical motion analyses of TMPRSS2 with and without binding with 1,25(OH)2D3 (correlated motion shown in read, anti-correlated motion shown in blue). The images were made with MATLAB (vR2015a, https://www.mathworks.com/products/matlab.html).
