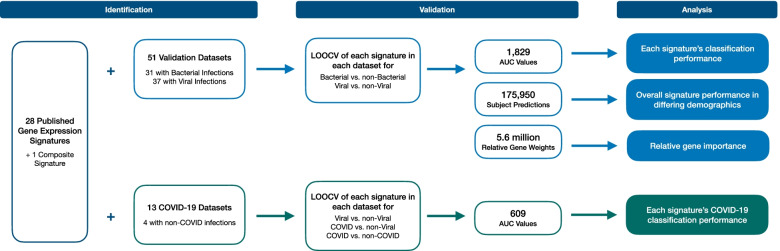
Fig. 1.
Study flow diagram. The performance of 28 published gene expression signatures and one composite signature was evaluated using leave-one-out cross-validation (LOOCV) in 51 publicly available datasets. LOOCV was performed for both bacterial vs. non-bacterial classification and viral vs. non-viral classification. LOOCV was also performed to measure the performance of signatures in 13 publicly available COVID-19 datasets. Performance was then measured by area under the receiving operating characteristic curve (AUC) values and individual subject predictions. Relative gene importance was characterized by the relative gene weights in each generated model