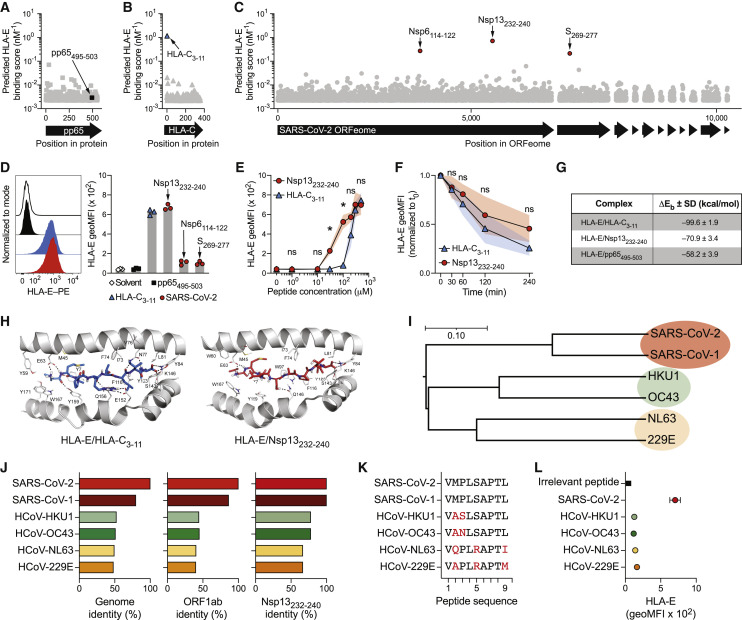
Figure 1.
SARS-CoV-2 Nsp13 encodes for an HLA-E-stabilizing peptide
(A–C) In silico prediction of HLA-E∗01:01-binding nonamers using NetMHC4.0. (A) pp65 protein containing the irrelevant pp65495–503 peptide. (B) HLA-C∗01:02 protein containing the known stabilizing HLA-C3–11 peptide. (C) SARS-CoV-2 ORFeome (isolate Wuhan-Hu-1) containing the three top candidates Nsp13232–240, Nsp6114–122, and S269–277.
(D) HLA-E stabilization on K562/HLA-E after pulsing with the indicated peptides at 300 μM overnight in serum-free medium. Left: representative HLA-E surface detection by flow cytometry (black line: solvent control; back filled histogram: pp65495-503; blue filled histogram: HLA-C3-11; red filled histogram: Nsp13232-240). Right: summary of HLA-E stabilization as geometric mean fluorescence intensity (geoMFI; n = 3 independent experiments).
(E) HLA-E surface stabilization after peptide pulsing with varying concentrations (n = 4 independent experiments).
(F) Pulse chase of HLA-E surface levels normalized to initiation of chase (t0; n = 4 independent experiments).
(G) Summary table of delta energy of binding as indicator of theoretical affinity of HLA-E/peptide complexes determined by molecular dynamics (MD) simulations (n = 3 replicate simulations).
(H) MD-based positioning of peptides in the peptide-binding groove of HLA-E. Left: HLA-C3–11. Right: Nsp13232–240.
(I) Phylogenetic relationships between the genomes of SARS-CoV-2, SARS-CoV-1, and common cold-causing HCoVs as determined by Clustalω. Scale bar indicates nucleotide substitution per site.
(J) Sequence identities relative to SARS-CoV-2. Left: genome level as determined by Clustalω. Middle: ORF1ab protein level as determined by Clustalω. Right: Nsp13232–240 peptide identity.
(K) Nsp13232–240 amino acid sequence comparison between viruses. Sequence alterations relative to SARS-CoV-2 are highlighted in red.
(L) HLA-E surface stabilization on K562/HLA-E after pulsing with Nsp13232–240 peptides from different viruses at 300 μM overnight determined by flow cytometry and displayed as geoMFI (n = 3 independent experiments).
Data are either mean and individual data points (D) or mean ± SD (E, F, G, and L). Statistical significance was tested using two-way repeated measures ANOVA with Bonferroni correction (E and F).
∗p < 0.05. See also Figure S1.