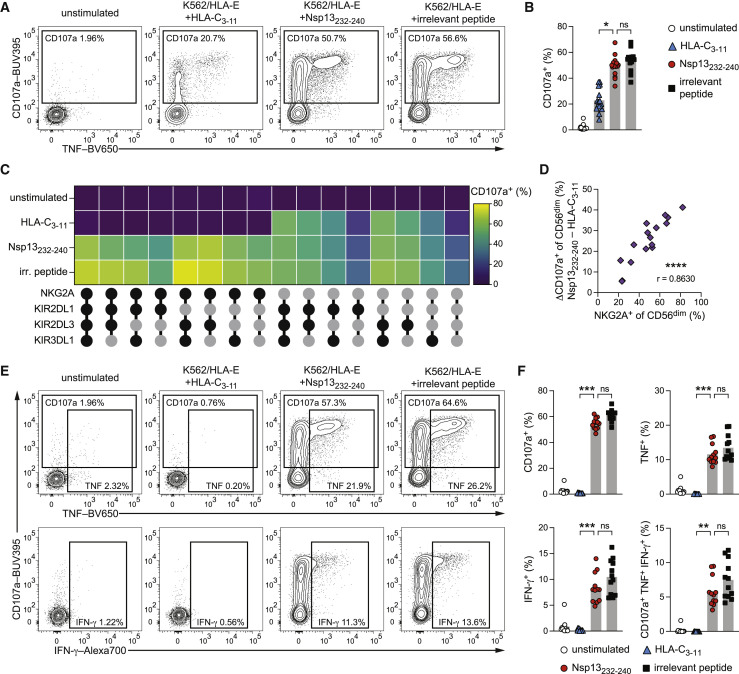
Figure 3.
Nsp13232–240 presented by HLA-E unleashes NKG2A+ NK cell activity
NK cell activation of purified NK cells from healthy donors either left unstimulated or stimulated by co-culture with peptide-pulsed K562/HLA-E target cells as determined by flow cytometry.
(A) Representative degranulation of CD56dim NK cells upon co-culture with K562/HLA-E cells pulsed with the indicated peptides as measured by CD107a surface mobilization detected by flow cytometry.
(B) Summary of degranulation (n = 16 donors in 6 independent experiments).
(C) Degranulation responses of CD56dim NKG2C− NK cell subsets stratified for expression of NKG2A, KIR2DL1, KIR2DL3, and KIR3DL1 (black filled circle: expressed; gray filled circle: not expressed) upon co-culture with K562/HLA-E cells pulsed with the indicated peptides (n = 8 donors in 4 independent experiments).
(D) Correlation of the frequency of NKG2A+ NK cells within the total CD56dim NK cells population and the difference in degranulation between pulsing with Nsp13232–240 and HLA-C3–11 (n = 16 donors in 6 independent experiments).
(E) Representative degranulation and intracellular cytokine expression of CD56dim NKG2C− NKG2A+ NK cells upon co-culture with K562/HLA-E cells pulsed with the indicated peptides.
(F) Summaries of activation of CD56dim NKG2C− NKG2A+ NK cells. Top left: degranulation. Top right: TNF expression. Bottom left: IFN-γ expression. Bottom right: polyfunctional (CD107a+ TNF+ IFN-γ+) responses (n = 12 donors in 5 independent experiments).
Data are either mean and individual data points (B and F), mean (C), or individual data points (D). Statistical significance was tested using Friedman test with Dunn's multiple comparison test (B and F) or Spearman correlation (D). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. irr., irrelevant. See also Figure S2.