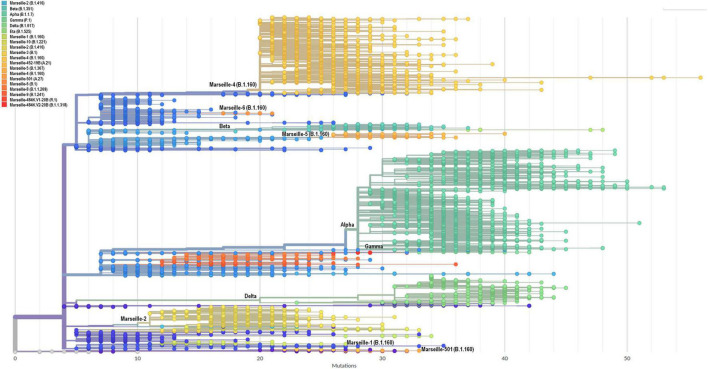
FIGURE 4.
Phylogenetic tree of 10,773 genomic sequences of SARS-CoV-2 obtained from patients SARS-CoV-2-diagnosed at IHU Méditerranée Infection, Marseille, France. Phylogeny reconstruction was performed using the nextstrain/ncov tool (https://github.com/nextstrain/ncov) then visualized with Auspice (https://docs.nextstrain.org/projects/auspice/en/stable/). The genome of the original Wuhan-Hu-1 coronavirus isolate (GenBank accession no. NC_045512.2) was added as outgroup. Major (most prevalent) variants are labeled. Marseille variants or WHO clades, and Pangolin, or Nextstrain clades are indicated. X-axis shows the number of mutations compared to the genome of the Wuhan-Hu-1 isolate (GenBank accession no. NC_045512.2).