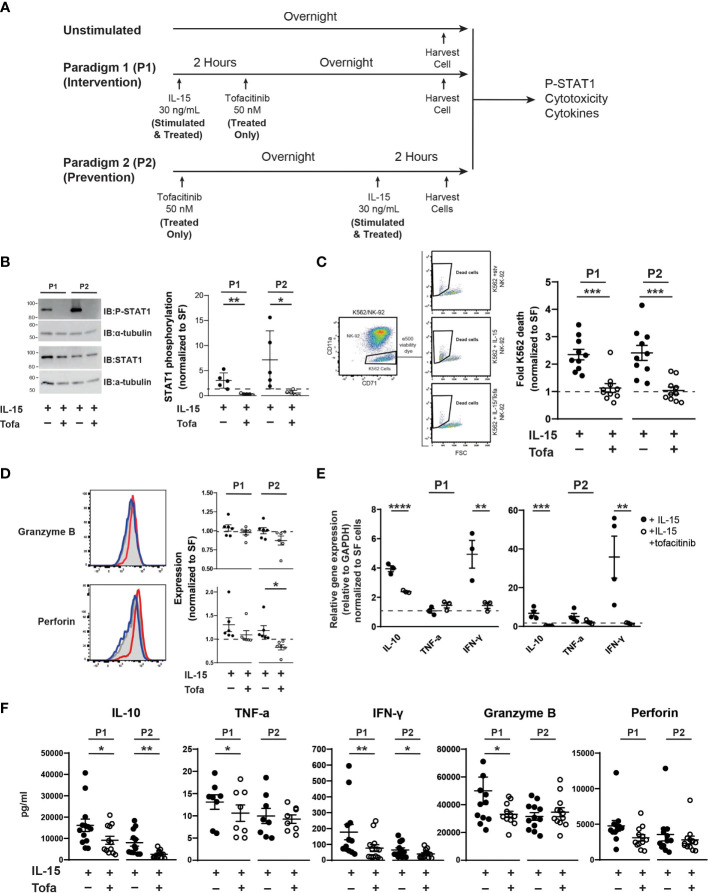
Figure 1.
Tofacitinib inhibits NK-92 cells function in vitro. (A) NK-92 cells were cultured with serum-free media under two culture paradigms: Paradigm 1 (P1) intervention treatment, whereby NK-92 cells were treated with IL-15 for two hours and then cultured overnight in the presence of tofacitinib (Treated), and Paradigm 2 (P2) prevention treatment where NK-92 cells were treated overnight with tofacitinib prior to two-hour IL-15 stimulation. (B) Stimulated and Treated NK-92 cells were assessed for STAT1 phosphorylation (P-STAT) following both culture paradigms. Representative immunoblots for Stimulated and Treated P-STAT1 and α-tubulin (internal reference) are shown. Quantitative data represent densitometric analysis, where total STAT1 and P-STAT1 signals were first normalized to α-tubulin then to P-STAT1 levels in Unstimulated cells; n = 5 independent experiments. (C) Unstimulated, Stimulated, and Treated NK-92 cells were co-cultured with K-562 cancer cells (10:1 ratio) following initial P1 or P2 culture paradigms. Flow cytometry was used to identify K-562 cells in the co-culture, and cell death was assessed by viability dye staining. K-562 cell death was quantitated for Stimulated and Treated NK-92 cells and normalized to Unstimulated NK-92 cells; n = 10 independent experiments. (D) Expression of the intracellular NK cell proteins perforin and granzyme B was determined by flow cytometry on Unstimulated, Stimulated, and Treated NK-92 cells using intracellular flow cytometry following P1 and P2 culture paradigms. Representative histograms are shown; gray peaks = Unstimulated NK-92 cells, red peaks = Stimulated NK-92 cells, blue peaks = Treated NK-92 cells show MFI. MFI from Stimulated and Treated cells were normalized to protein levels in Unstimulated NK-92 cells for quantitation; n = 6 independent experiments. (E) Gene expression of IL-10, TNF-α, and IFNγ cytokines was assessed in Unstimulated, Stimulated, and Treated NK-92 cells following P1 and P2 paradigms using qRT-PCR. Data for Stimulated and Treated cells were normalized to GAPDH expression then to the Unstimulated cells; n = 3-4 independent experiments. (F) Extracellular expression of IL-10, TNF-α, IFN-γ, granzyme B, and perforin was assessed for Stimulated and Treated NK-92 cells using a CD8/NK cell multiplex analysis following P1 and P2 treatment paradigms (n = 8-13 independent experiments). For all experiments, quantitative data is shown as the mean ± SEM; (A–E) comparisons were made by Student’s t-test; (F) comparisons were made using a paired t-test or a Wilcoxon test based on normality of the data. Horizontal dashed lines represent normalized Unstimulated NK-92 cell levels. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.