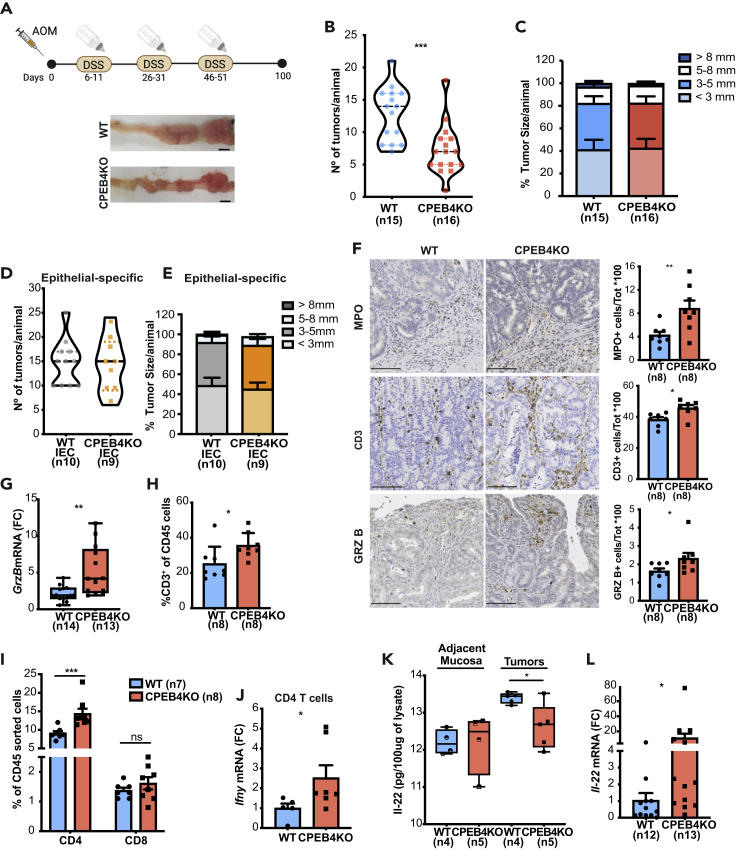
Figure 5.
CPEB4 deficiency reduces CAC development and shapes adaptive tumoral immune response, controlling IL-22 production
(A) Schematic representation of the two-stage murine model of CAC. Mice were intraperitoneally injected with one dose of azoxymethane (AOM, tumor-inducing agent), followed by three rounds of 2.5% dextran sodium sulfate (DSS, tumor-promoting agent) in drinking water. Representative tumor images in WT and CPEB4KO mice. Scale bar, 5 mm.
(B and C) Tumor number per animal (B) (WT, n = 15; CPEB4KO, n = 16; mean ± SEM; Mann-Whitney test, ∗∗∗p = 0.0006) and percentage of tumor sizes (C) in mm.
(D and E) Average tumor number (D) and size (E) of Villin-CreERT2 Cpeb4+/+(IEC WT, n = 10) and Villin-CreERT2 Cpeb4lox/lox (IEC CPEB4KO, n = 9) mice. Data represent mean ±SEM.
(F) Representative stainings of myeloperoxidase (MPO, an enzyme produced by neutrophil granulocytes to potentiate inflammation and apoptotic oxidative damage), T lymphocytes (CD3) and granzyme B (GrzB) in AOM/DSS colon tumor sections from WT and CPEB4KO mice (n = 8/genotype). Related quantifications are shown in the histograms. Data represent means ±SEM ∗p = 0.0111; ∗∗∗p = 0.0003; ∗p = 0.0207 (unpaired t-test for MPO; Mann-Whitney test for CD3 and GrzB).
(G) Granzyme B mRNA levels, relative to Gapdh, in colon tumors from WT (n = 14) and CPEB4KO (n = 13) mice. ∗∗p = 0.0023 (Mann-Whitney test).
(H) Flow cytometry analysis of CD3+ T lymphocytes (gated from CD45+ cells) from WT and CPEB4KO mice. Data are mean ± SEM (n = 7/genotype). ∗p = 0.0148 (Mann-Whitney test).
(I) Flow cytometry analysis of CD4 and CD8 T lymphocyte populations of colon tumors from WT (n = 7) and CPEB4KO (n = 8) mice. ∗∗∗p = 0.0001 (two-way ANOVA test, multiple comparisons).
(J) Ifnγ mRNA levels, relative to Gapdh, in CD4 tumor sorted T cells from WT (n = 5) and CPEB4KO (n = 7) mice. ∗p = 0.048 (Mann-Whitney test).
(K) IL-22 ELISA in adjacent mucosa and AOM-DSS tumors of WT (n = 4) and CPEB4KO (n = 5) mice. ∗p = 0.0241 (unpaired t-test).
(L) Relative Il-22 mRNA levels in WT (n = 12) and CPEB4KO (n = 13) tumors analyzed by RT-qPCR ∗p = 0.0109 (unpaired t-test). Data are pooled from two (B,C,G,L) or representative of two (D,E) biologically independent experiments