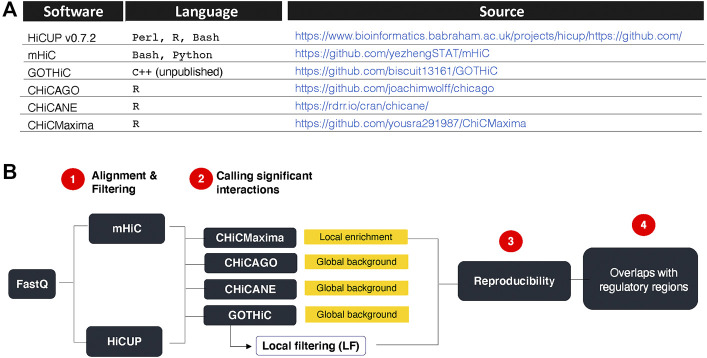
FIGURE 1.
Research summary. (A). CHi-C analytical tools used and their sources. (B). Strategy overview. HiCUP and mHiC were compared for their performance in mapping read pairs and filtering experimental artefacts. GOTHiC, CHiCMaxima, CHiCAGO and CHiCANE were compared for their ability to identify reproducible, biologically relevant interactions. Yellow boxes indicate the type of background model used by each tool. GOTHiC local filtering (LF) is an optional downstream filtering of GOTHiC globally significant interactions based on the local interaction profile of each bait.