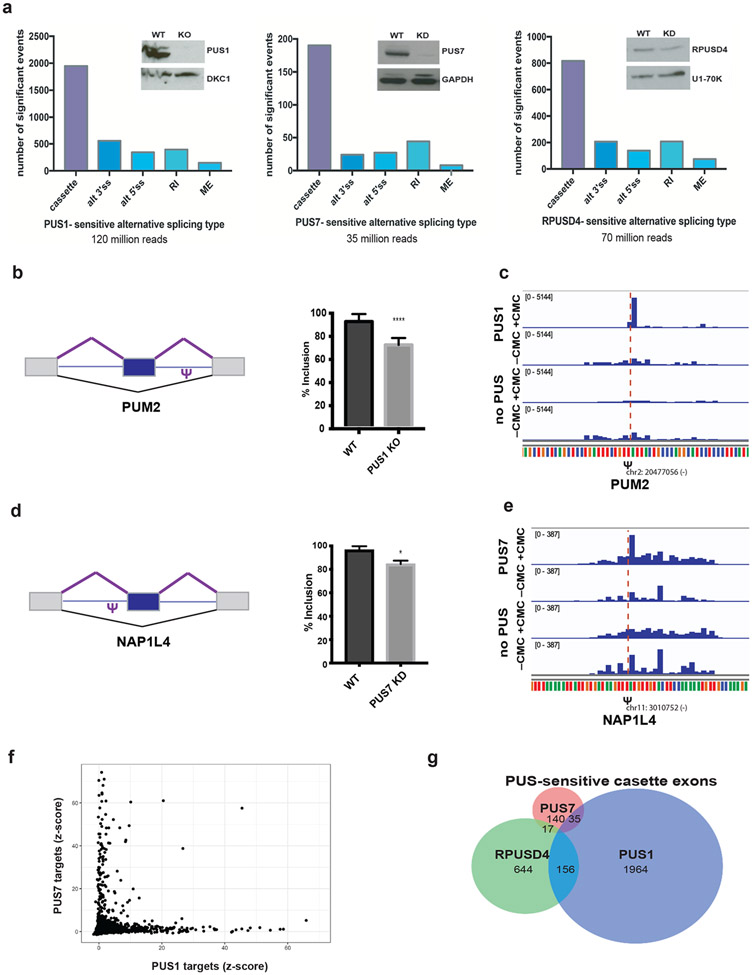
Figure 5. Pseudouridine synthases regulate alternative splicing.
a) Left - Western blot of the CRISPR knockout PUS1 HepG2 cell line probed for PUS1 and a loading control. RNA was isolated from PUS1 knockout (KO) and wild type (WT) cells and mRNA-seq libraries were prepared from poly(A)+ mRNA. The number of significant alternative splicing changes in PUS1 KO versus WT (n=2 biological replicates) is displayed by type of alternative splicing: cassette exons (cassette), alternative 3′ splice sites (alt 3′ss), alternative 5′ splice sites (alt 5′ ss), retained introns (RI) and mutually exclusive exons (ME). Significant alternative splicing events were determined from rMATS as those events that changed by greater than 10% difference in percent inclusion and a false discovery rate (FDR) of less than or equal to 0.05. Middle - Western blot of representative RPUSD4 knockdown (~60%) at 96h following shRNA induction. RPUSD4-sensitive alternative splicing changes determined from RNA-seq analysis (n=2 biological replicates) as above. Right - Western blot of representative PUS7 knockdown (~90%) at 96h following shRNA induction. PUS7-sensitive alternative splicing changes determined from RNA-seq analysis (n=3 biological replicates) as above. b) Left - schematic of a cassette exon in PUM2 and location of pseudouridine. Right - quantification of exon inclusion in WT and PUS1 KO based on junction spanning reads from RNA-seq. Asterisk denotes statistical significance based on p-value < 0.05 as calculated by rMATS. c) Genome browser view of Pseudo-seq reads of the intronic PUM2 pseudouridine site (Figure 5b) following pseudouridylation with recombinant PUS1 or in the absence of PUS. d) Left - schematic of a cassette exon in NAP1L4 and location of pseudouridine. Right - quantification of exon inclusion in WT and PUS7 KD based on junction spanning reads from RNA-seq. Asterisk denotes statistical significance based on p-value < 0.05 as calculated by rMATS. e) Genome browser view of Pseudo-seq reads of the intronic NAP1L4 pseudouridine site (Figure 5d) following pseudouridylation with recombinant PUS7 or in the absence of PUS. f) Scatter plot showing pairwise comparisons of Z-score values at candidate pseudouridine sites incubated with recombinant PUS1 versus PUS7. g) Venn diagram of overlap among cassette exons regulated by PUS1, PUS7 and RPUSD4.