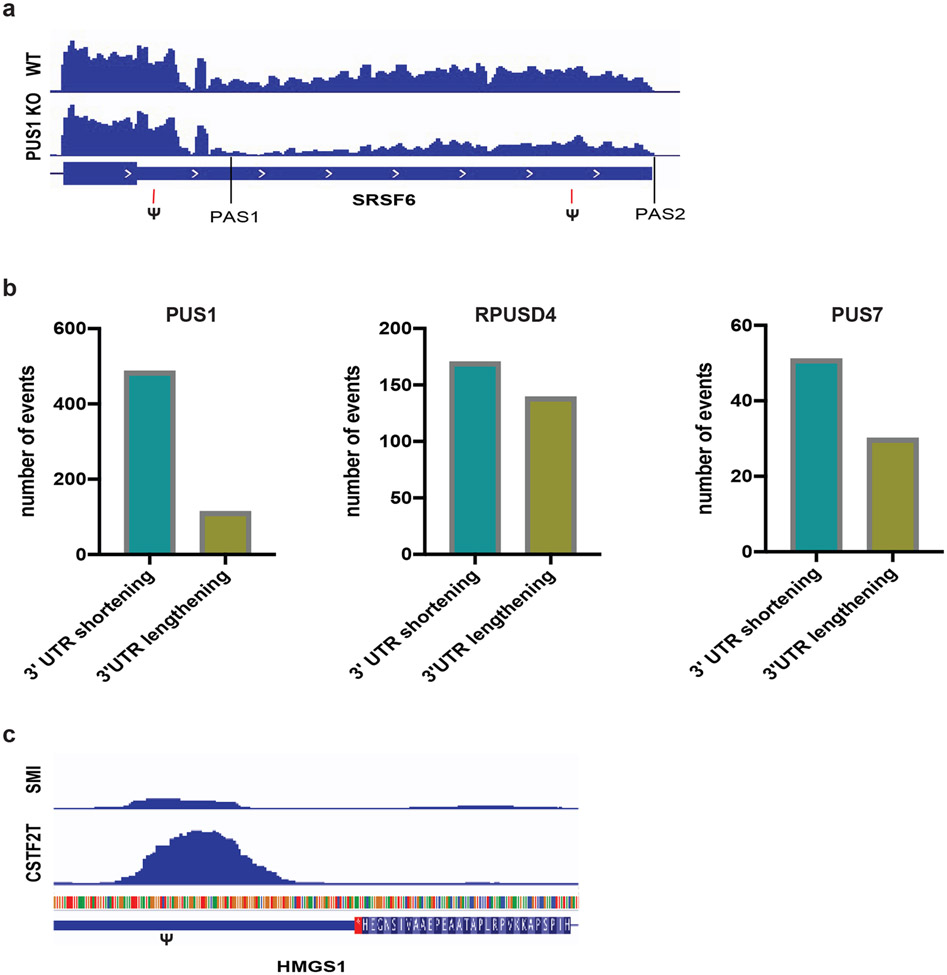
Figure 6. Pseudouridine synthases regulate 3′ end processing.
a) Genome browser view of a PUS1-dependent alternative cleavage and polyadenylation (APA) in the 3′ UTR of SRSF6. Upon PUS1 KO there is a shift toward usage of a proximal polyA site (PAS1) and away from the diatal polyA site (PAS2) resulting in expression of a shorter 3′ UTR isoform. The location of a two pseudouridine in this 3′ UTR upstream of each PAS is indicated. b) The number of significant alternative cleavage and polyadenylation events in PUS1 KO (n=2 biological replicates), RPUSD4 KD (n=2 biological replicates) or PUS7 KD (n=3 biological replicates) compared to WT is displayed by type of APA: 3′ UTR shortening and 3′ UTR lengthening. Significant APA events were determined from QAPA as those events that changed by greater than 10% difference in polyA site usage and were reproducible across replicates. c) Genome browser views of CSTF2T eCLIP peak and size-matched input controls (SMI) in the 3′ UTR of HMGS1. The location of a pseudouridine relative to the eCLIP peak is denoted by (Ψ).