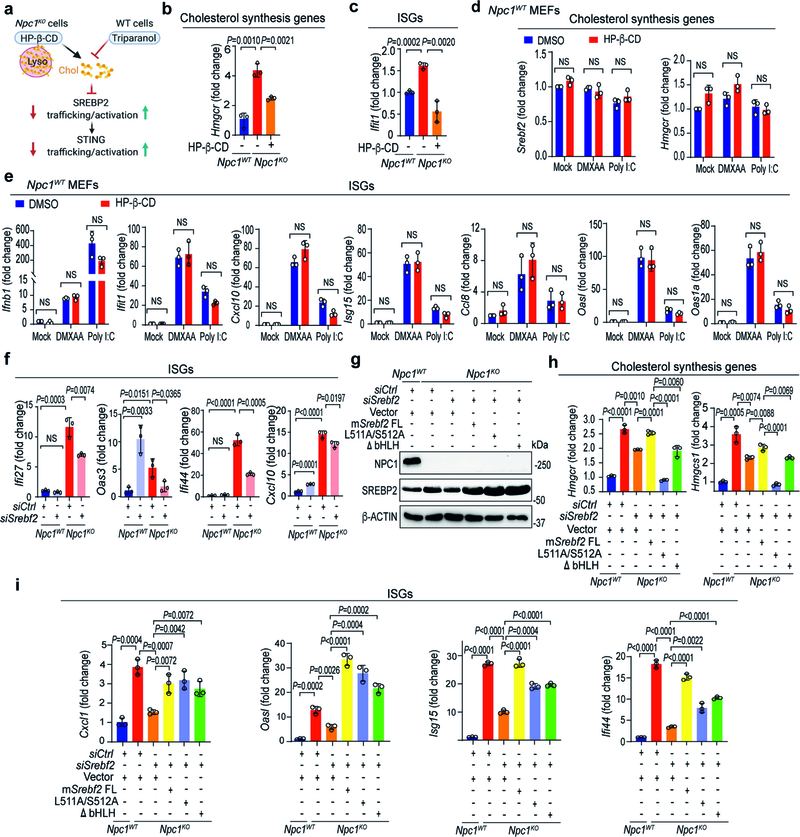
Extended Data Fig. 3 |. SREBP2 trafficking primes STING signalling in Npc1KO cells independently of its transcriptional activity.
a, Diagram showing mechanisms of action for HP-β-CD and triparanol on cholesterol synthesis and SREBP2 activation. HP-β-CD promotes the egress of lysosomal cholesterol to the ER, thus limiting SREBP2 trafficking and activation in Npc1KO cells. Triparanol inhibits the biochemical conversion of desmosterol into cholesterol, thus promoting SREBP2 trafficking and activation in wild-type cells. b, c, qRT–PCR analysis of cholesterol-synthesis genes (b) and ISGs (c) in Npc1WT and Npc1KO MEFs mock-treated or treated with HP-β-CD (0.3 mM) overnight (n = 3). d, e, qRT–PCR analysis of mock-, DMXAA- (50 μg ml−1, 2 h) or poly(I:C)- (1 μg ml−1, 2 h) induced expression of cholesterol-synthesis genes (d) and ISGs (e) with mock- or HP-β-CD treatment (0.3 mM, 8 h) in Npc1WT MEFs (n = 3). f, qRT–PCR analysis of the baseline expression of ISGs in Npc1WT and Npc1KO MEFs transfected with siCtrl or siSrebf2 for 48 h (n = 3). g, Immunoblot analysis of the indicated proteins (left) in Npc1WT, Npc1KO, Npc1KOSrebf2KD and Npc1KOSrebf2KD MEFs reconstituted with SREBP2 wild type (FL) or transcription-inactive mutants (L511A/S512A, ΔbHLH). h, i, qRT–PCR analysis of the expression of cholesterol-synthesis genes (h) and ISGs (i) in Npc1WT, Npc1KO, Npc1KOSrebf2KD and Npc1KOSrebf2KD MEFs reconstituted with SREBP2 wild type (FL) or transcription-inactive mutants (L511A/S512A, ΔbHLH) (n = 3). b–f, h, i, Data are mean ± s.d. Unpaired two-tailed Student’s t-test. NS, not significant. Data are representative of at least two independent experiments.