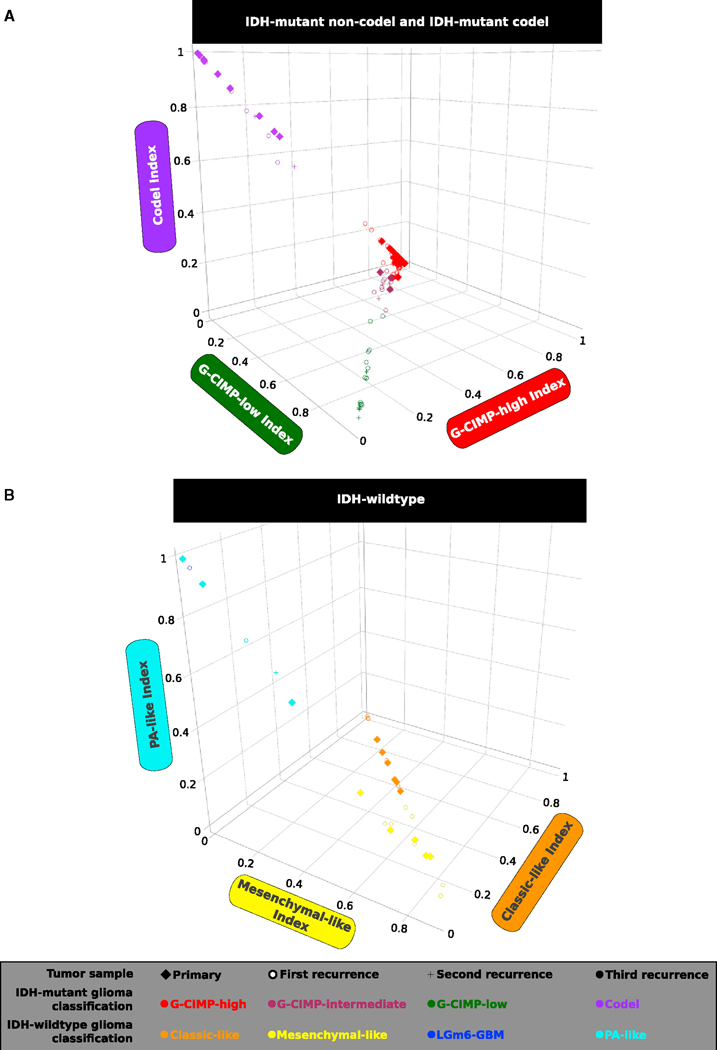
Figure 1. Identification of Longitudinal Tumors with a G-CIMP-High to G-CIMP-Low Epigenetics Shift during Recurrence and Malignant Tumor Progression.
The methylomes of 200 longitudinally collected TCGA and non-TCGA adult diffuse gliomas (grades II to IV) from 77 patients profiled on the 450,000 platform were classified by supervised random forest (RF) computational approaches into one of the 7 pan-glioma DNA methylation subtypes (accuracy > 95% on average) using the CpG probe signatures described in Ceccarelli et al. (2016).
(A) This 3D scatterplot using IDH mutant Codel (negative control of G-CIMP signatures) and IDH mutant non-Codel G-CIMP-high and G-CIMP-low indices predicted by the RF model shows a distinct set of samples within the IDH mutant non-Codel G-CIMP subtypes exhibiting relatively intermediate DNA methylation profiles. This subgroup of samples has been named G-CIMP-intermediate post-RF assessment. A subset of initially LGG G-CIMP-high tumors switches to a G-CIMP-low phenotype at first recurrence, whereas a subset of tumors retains their original G-CIMP-high phenotype at first recurrence as a form of epigenetic memory.
(B) 3D scatterplot using IDH-wild-type PA-like, classic-like, and mesenchymal-like indices predicted by RF shows that IDH-wild-type gliomas do not change significantly in terms of their DNA methylation patterns during disease relapse.