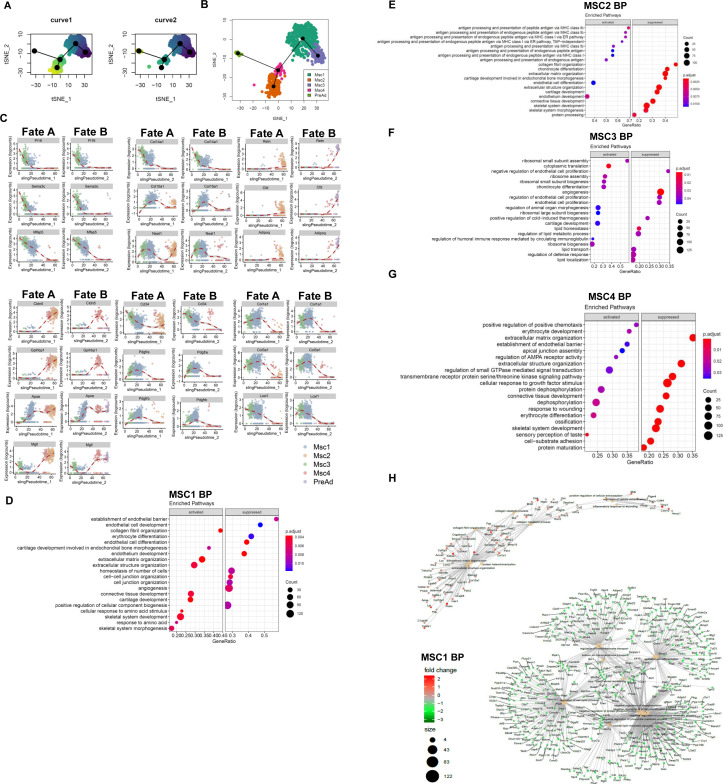
Figure S2. Progenitor cells produce ECM.
(A) Pseudotime trajectory curve for each lineage (B) t-SNE of inferred trajectory of progenitor cells and preadipocytes. (C) Expression profiles of select genes during both lineages identified; dots represent a cell and line represents the loess regression. (D, E, F, G) Dot plot of gene ontology (molecular function) of activated and suppressed pathways in (D) MSC1, (E) MSC2, (F) MSC3, (G) MSC4 when compared with each other. (H) Gene concept network showing enriched terms for the genes down-regulated or up-regulated by high-fat diet feeding in MSC1 population; the color of the nodes representing log2 fold change in gene expression relative to the low-fat diet MSC1.