Figure 1.
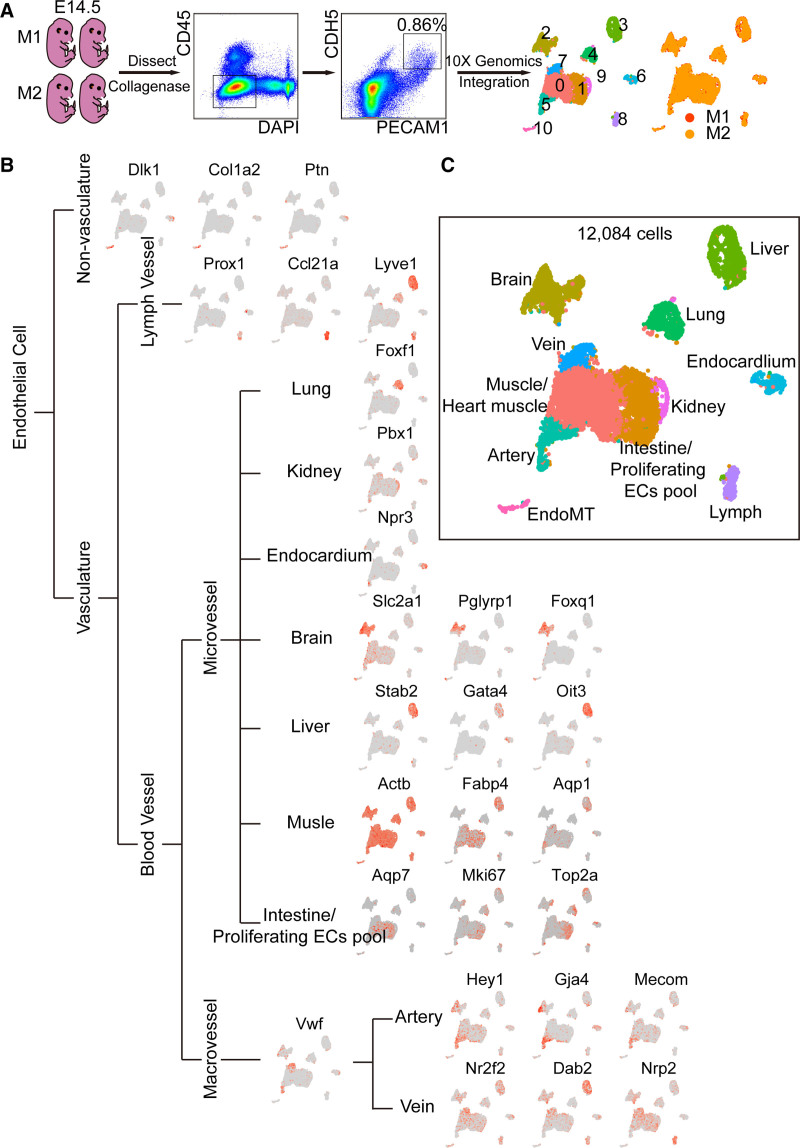
Endothelial cells (ECs) at embryonic day 14.5 were divided into 11 populations. A, ECs were enriched for single-cell RNA sequencing analysis. Collagenase I was used to dissect embryos at day 14.5 (E14.5). Fluorescence-activated cell sorting (FACS) was used to isolate live (DAPI−), nonmonocyte (CD45−) ECs (PECAM1 [CD31+] and CDH5+). Seurat 3.0 was used for single-cell analysis. Colors denoted different clusters or different origins of mouse. B, Known endothelial mark genes in different tissues were used to define clusters of embryonic ECs (eECs). C, eECs were classified into 11 populations. Colors denoted different populations. DAPI indicates 4',6-diamidino-2-phenylindole; EndoMT, endothelial-to-mesenchymal transition; M1, embryos from mouse 1; M2, embryos from mouse 2; and PECAM1, platelet endothelial cell adhesion molecule-1.