Figure 3: Gain-of-function phenotype of UM-associated PLCβ4 mutations and the importance of the negatively charged Asp630 to PLCβ4 activity.
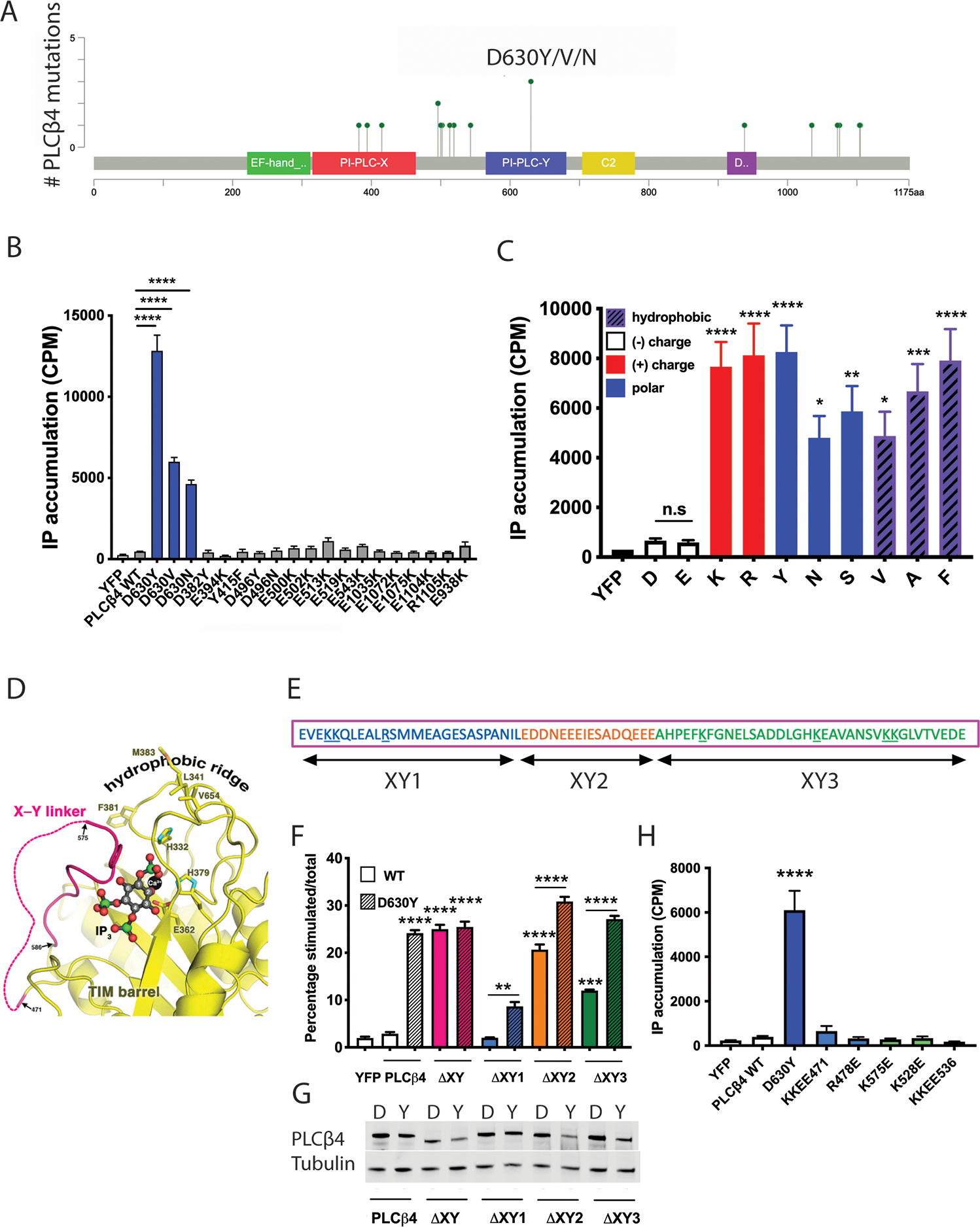
(A) A lollipop plot represents the positions of PLCβ4 mutations found in UM (from the TCGA database) or other cancers (COSMIC database). (B) Assessment of the activity of UM-associated PLCβ4 mutants D630Y/V/N and other charge-altering mutants by transfection of these mutant constructs to COS-7 cells. [3H]IP accumulation was assayed as in Fig. 1C. (C) Effect of replacement of the negative charge of Asp (or Gly) at residue 630 by other amino acids on PLCβ4 activity, assessed in COS-7 cells by measuring total [3H]IP accumulation. (D) Modeling of IP3 (derived from PDB ID 1DJX) bound to the PLCβ3 active site, wherein the X-Y linker forms a lid to occlude substrate from accessing the catalytic site. (E) Linear schematic of the X-Y linker in PLCβ, consisting of 3 sub-regions comprised of a disordered region at the amino terminal (blue; XY1), an acidic stretch region (orange; XY2), and a helix at the carboxyl terminal (green; XY3). Positively charged residues are underlined. (F) [3H]IP accumulation was measured in COS-7 cells transfected with mutants of PLCβ4, in which the whole or sub-regions of the X-Y linker were deleted in the WT (“D”) or D→Y mutant (“Y”) PLCβ4 background. (G) Cropped Western blotting for the abundance of the X-Y linker mutants in the WT (“D”) or D→Y mutant (“Y”) PLCβ4 background in (F). (H) ([3H]IP accumulation measurement in COS-7 cells transfected with PLCβ4 point-mutation constructs, in which positively charged residues in the X-Y linker were mutated to negatively charged glutamate residues in the WT (“D”) PLCβ4 background. In all panels, data are representative or are mean ± SD from at least three independent experiments. * P < 0.05, ** P < 0.01, *** P < 0.001, and **** P < 0.0001 by one-way ANOVA with Bonferroni post-test; where not otherwise indicated with brackets, comparison was with WT (“D”) PLCβ4.
