Figure 3.
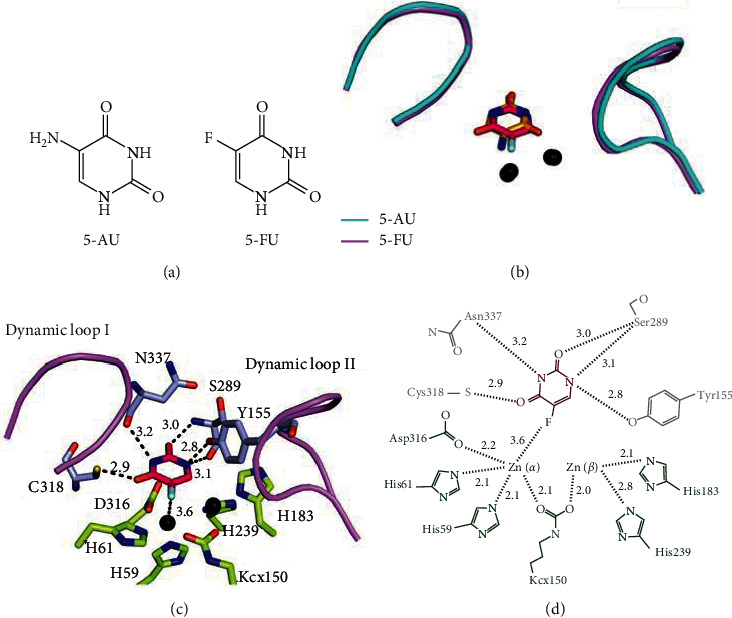
Comparison between the 5-AU and 5-FU binding modes. (a) The structure of 5-AU and 5-FU. (b) The superimposed structures of the 5-AU and 5-FU bound states. The conformation of the dynamic loops in both binding states is similar, but the orientations between 5-AU and 5-FU (hot pink) are different. (c) The interactions of PaDHPase with 5-FU. Znα and the substrate binding sites (gray) in PaDHPase were involved in 5-FU binding. Dynamic loops are colored in light pink. 5-FU interacted with the main chains of residues Ser289 and Asn337 and the side chains of residues Tyr155 and Cys318. The distances are shown on dotted lines. (d) 5-FU interacted with Znα, Ser289, Cys318, Asn337, and Tyr155. Residues (His59, His61, Kcx150, His183, His239, and Asp316) required for metal binding are colored in green.
