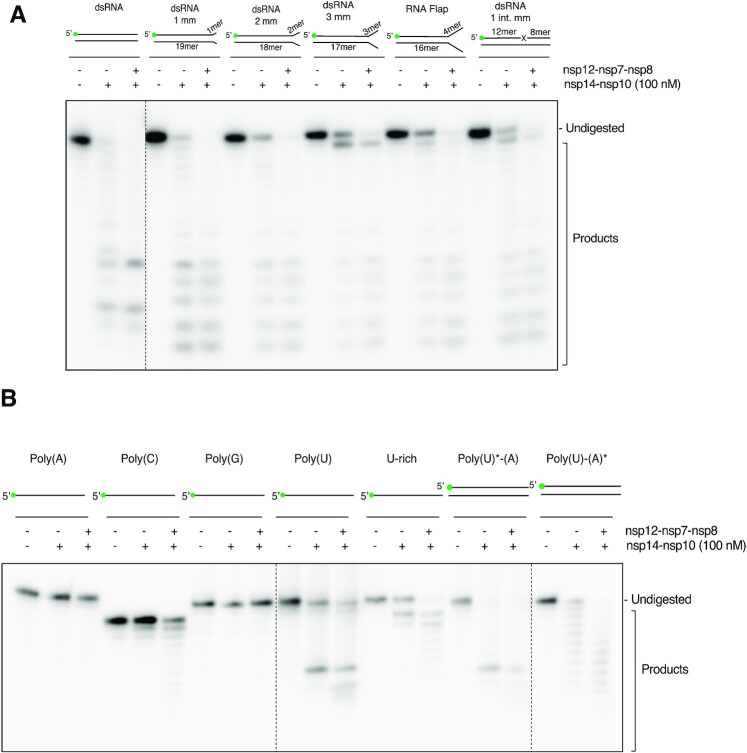
Figure 3.
The SARS-CoV-2 polymerase nsp12-nsp7-nsp8 complex enhances nsp14–nsp10 nuclease activity on a variety of RNA substrates.(A) In the presence of the nsp12–7–8 polymerase complex, nsp14–nsp10 shows enhanced activity on a variety of RNA substrates, including RNA substrates with mismatched termini and flaps with no observed differential effects. mm: mismatch int mm: internal mismatch. B. When presented with 20-mer Poly(A), Poly(C), Poly(G), Poly(U) and U-rich ssRNA (oligonucleotides 12, 13, 14, 11 and 25 respectively, see Supplementary Table 1A) as well as double-stranded Poly(U)*-(A) and Poly(U)-(A)*, nsp14–nsp10 shows a more profound stimulation on repetitive sequences when nsp12–7–8 is present. Enhancement of nsp14–nsp10 nuclease activity by nsp12–7–8 is most pronounced on uracil-containing substrates. * Indicates labelled strand. The SARS-CoV-2 nsp12–7–8 polymerase complex (at a 1:3:3 molar ratio) was incubated with 10 nM substrate (37 °C, 5 min) prior to addition of 100 nM of nsp14–nsp10 at a 1:2 molar ratio of nsp14–nsp10 complex to nsp12–7–8 complex. Reactions were incubated at 37 °C for 45 min, then analysed by 20% denaturing PAGE. The size of products was determined as shown in Supplementary Figure 1E. Main products are labelled *, ** and *** corresponding to 12-mer, 10-mer and 7-mer respectively. Oligonucleotides used are given in Supplementary Table 1A and B.