Figure 4.
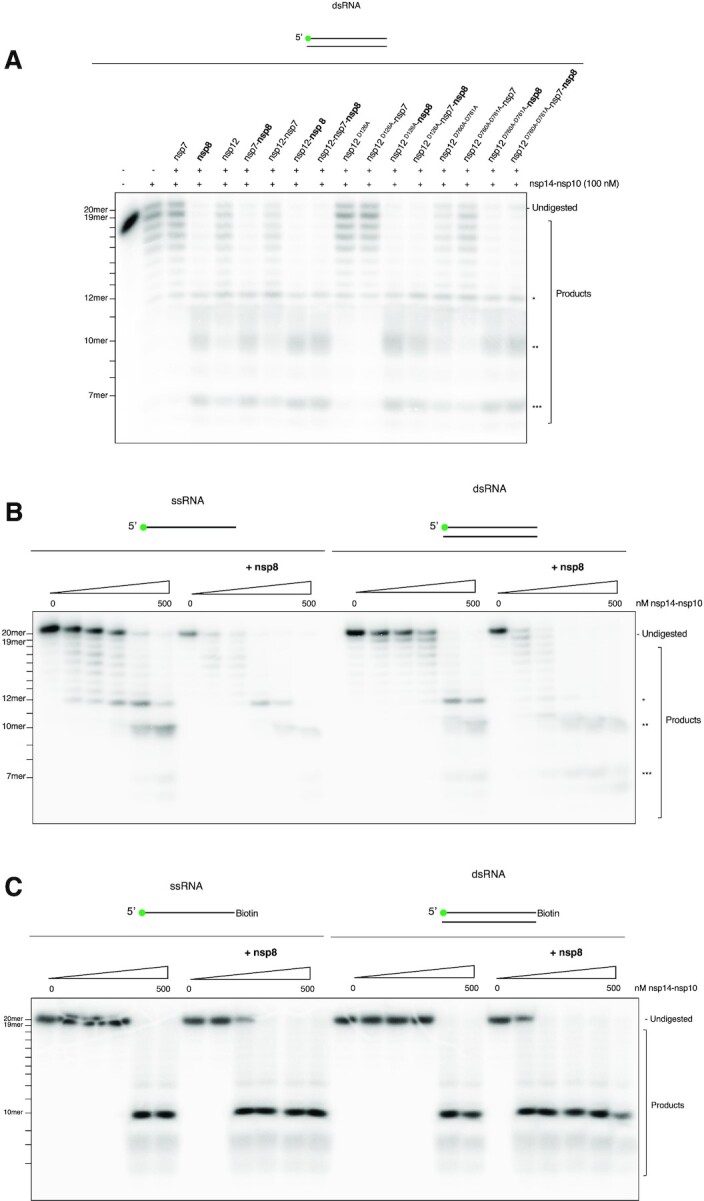
The SARS-CoV-2 polymerase nsp12–nsp7–nsp8 complex enhances nsp14–nsp10 nuclease activity on a variety of RNA substrates.(A) The SARS-CoV-2 primase, nsp8 enhances nsp14–nsp10 activity. The addition of each individual polymerase subunit, i.e.nsp12, nsp7, and/or nsp8, as well as different combinations reveal nsp8 as the major enhancer of nsp14–nsp10 activity. Mutant nsp12D126A and nsp12D760A-D761A show no substantial stimulation of activity of nsp14–nsp10 compared to nsp8 suggesting stimulation of nuclease activity is uncoupled from polymerase activity. (B) The presence of nsp8 stimulates the exonuclease activity of SARS-CoV-2 nsp14–nsp10. Increasing concentrations of nsp14–nsp10 were incubated with ss- or dsRNA (Oligo 2 and 3, see Supplementary Table 1A) and a fixed concentration of 300 nM nsp8 (37°C, 45 min). Reactions were analysed by 20% denaturing PAGE. (C) The presence of nsp8 stimulates the endonuclease activity of nsp14–nsp10. Increasing concentrations of nsp14–nsp10 were incubated with ss- or dsRNA with a blocked (3′-biotin) terminus (Oligo 2 and 16 see Supplementary Table 1A) and a fixed concentration of 300 nM nsp8 at (37°C, 45 min). Reactions were analysed by 20% denaturing PAGE. The SARS-CoV-2 nsp12–7–8 polymerase complex (at a 1:3:3 molar ratio) was incubated with 10 nM substrate (37°C, 5 min), prior to addition of 100 nM of nsp14–nsp10 at a 1:2 molar ratio of nsp14–nsp10 complex to nsp12–7–8 complex. Reactions were incubated at 37°C for 45 min and subsequently analysed by 20% denaturing PAGE. The size of products was determined as shown in Supplementary Figure 1E. Main products are labelled *, ** and *** corresponding to 12-mer, 10-mer and 7-mer respectively. Oligonucleotides used are given in Supplementary Table 1A and B.
