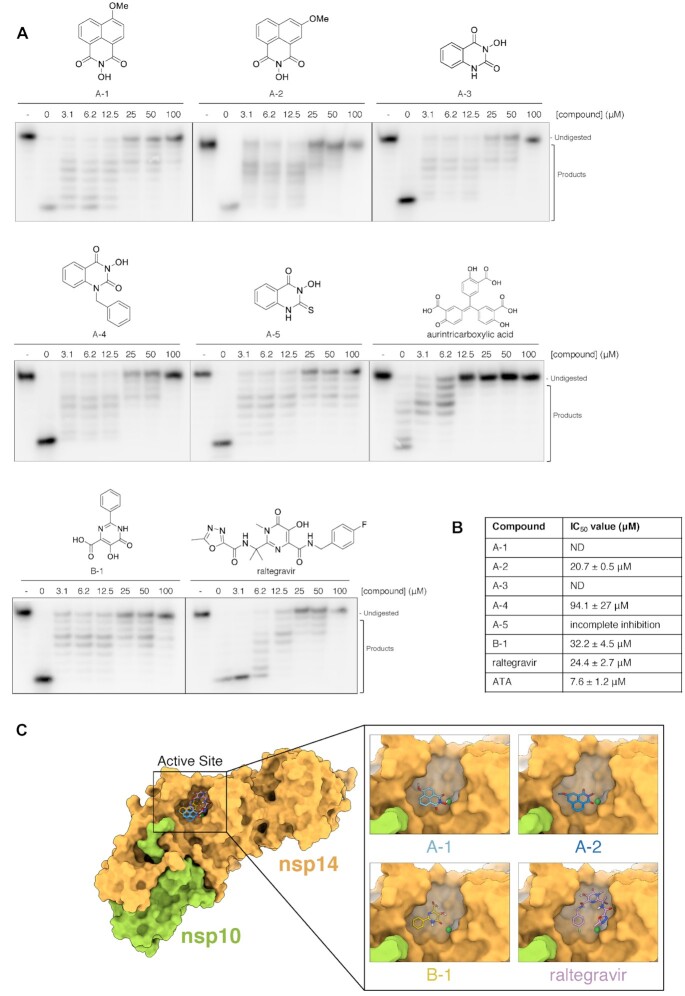
Figure 5.
The exonuclease activity of nsp14–nsp10 is inhibited by N-hydroxyimide and hydroxypyrimidinone based compounds. (A) Increasing concentrations (as indicated, in μM) of compounds incubated with 100 nM nsp14–nsp10 (room temperature, 10 min), before initiating nuclease reaction by addition of ssRNA (37 °C, 45 min). Products were analysed by 20% denaturing PAGE. A decrease in the generation of nucleolytic reaction products and an increase in undigested substrate indicates inhibition of nuclease activity at increasing inhibitor concentrations. - indicates no enzyme. Compounds A-1–A-4 are based on a N-hydroxyimide scaffold, B-1 is a hydroxypyrimidinone. (B) IC50 values determined by quantification of gel digestion products (100 nM nsp14–nsp10); dose-response curves were determined by nonlinear regression. The mean ± s.e.m. were calculated from ≥3 biological repeats. (C) Docking of nsp14–nsp10 using Autodock. Nsp14–nsp10 was docked with compounds within grid boxes encompassing a surface focussed on the active site surface then the highest-affinity docking pose of A-1 and A-2 overlaid on the surface of SARS-CoV nsp14–nsp10; Mg2+ is in dark green and the highest-affinity docking poses of B1 and raltegravir overlaid on the surface of SARS-CoV nsp14–nsp10. Nsp14 is in yellow-orange, nsp10 is in light green, Mg2+ is in dark green. The docked poses of the compounds on the surface of the whole nsp14–nsp10 complex are shown on the left-hand side, with a detailed view at the active site on the right-hand side inset.