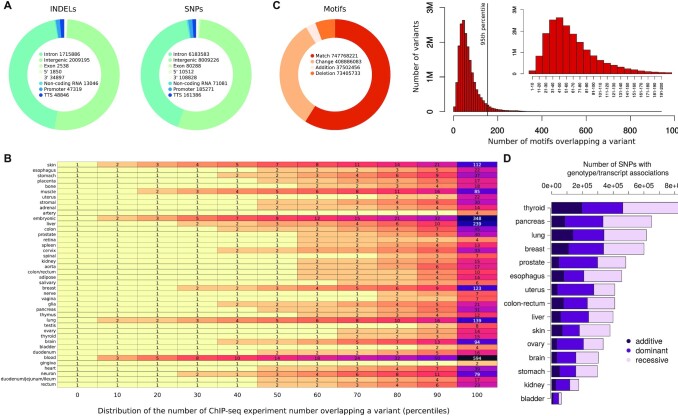
Figure 2.
Summary of the data contained in Polympact. (A) Annotations for the different types of variants stored in Polympact. (B) Percentiles of the distribution of the number of variants overlapping a ChIP-seq peak in various tissues. (C) Types of binding motifs results and distribution of the number of motifs overlapping a variant for match, change, addition and deletion types. In small, a zoom of the major distribution part. (D) Number of variants associated with a transcript level in additive, dominant and recessive models in different tissues.