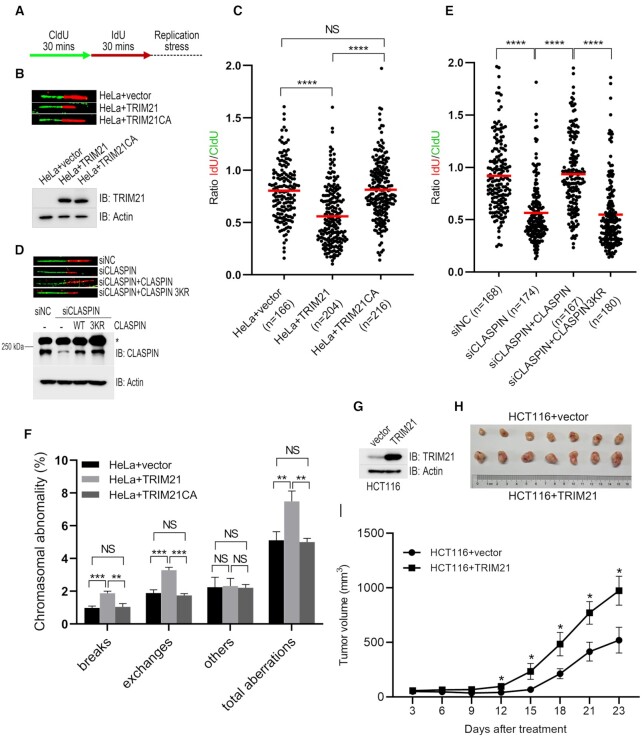
Figure 6.
Overexpression of TRIM21 led to genome instability and tumorigenesis. (A) Schematic of the DNA fiber assay examining stalled replication fork stability. Cells were sequentially labeled with 40 μM CldU and 100 μM IdU for 30 min each, followed by treatment with 5 mM hydroxyurea and 5 μM aphidicolin to block replication fork progression. (B, C) HeLa cells or HeLa cells overexpressing either TRIM21 or TRIM21(CA) were treated as described in (A); the DNA fibers were spread and subjected to immunofluorescence using anti-BrdU antibodies specifically recognizing CldU or IdU. (B) Representative images of the DNA fibers and the TRIM21 expression level. (C) For each group, a IdU/CldU ratio of > 150 individual DNA fibers is presented and the mean IdU/CldU ratio (marked as the red line) is shown. ****, t-test, P < 0.0001. (D, E) HeLa cells and HeLa cells expressing exogenous CLASPIN or ubiquitination-defective mutant CLASPIN(K565/580/581R) were transfected with siNC or siCLASPIN for 48 h before subjected to DNA fiber assay as described in (A) and immunoblotting with antibodies as indicated. Representative images of the DNA fibers and the CLASPIN expression levels are shown in (D). * Non-specific signal. (E) For each group, a IdU/CldU ratio of >150 individual DNA fibers is presented and the mean IdU/CldU ratio (marked as the red line) are shown in (E). ****, t-test, P < 0.0001. (F) HeLa cells stably overexpressing TRIM21 or its E3 ligase inactive mutant were subjected to metaphase spread assay; >1500 metaphase chromosomes were analyzed for every cell line. The error bars represent the SD, n = 3. t-test, **P < 0.01; ***P < 0.001. (G–I) 6 × 106 HCT116 cells overexpressing empty vector or TRIM21 were implanted into nude mice, and the tumor volume was monitored at the indicated times. The TRIM21 expression level was examined (G). Tumor images (H) and quantification results are shown (I). n = 7, mean tumor volume ± SEM, t-test, *P < 0.05.