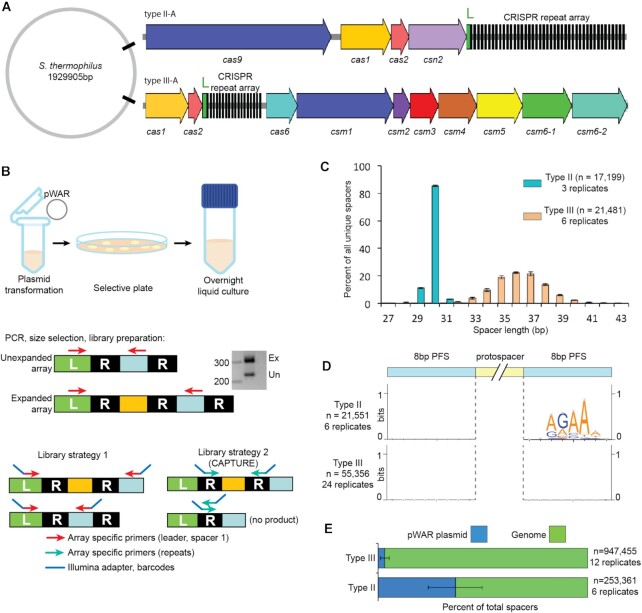
Figure 1.
Adaptation by type III-A and type II-A CRISPR-Cas systems. (A) The schematic of CRISPR-Cas systems of Sth JIM8232. The cas genes are represented by colored arrows, ‘L’ indicates the leader sequences in green, and the repeats of the CRISPR array are shown in black. (B) Schematic showing steps in the adaptation assay. After isolation of genomic DNA, PCR is used to amplify the leader-adjacent end of the CRISPR array. Expanded PCR products (Ex) contain a new leader-adjacent repeat-spacer unit and so are approximately 70 bp longer; unexpanded PCR products (Un) reflect the original, WT CRISPR array. (C) Length distribution of the new spacers acquired into the type II-A system (cyan) and the type III-A system (peach). (D) Analysis of protospacer adjacent sequences (PFS). Sequences upstream and downstream from aligned new spacers were extracted and used to create weblogos to visualize sequence motifs. (E) Proportion of new spacers that aligned to the plasmid (blue) and genome (green). Pooled data of at least three independent experiments are presented in (C–E).