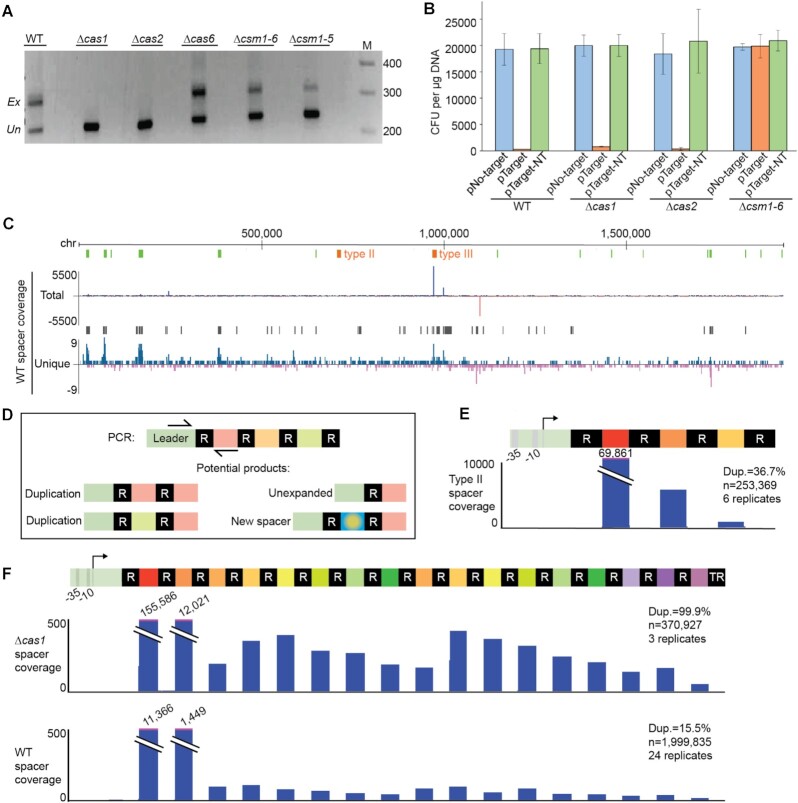
Figure 2.
Cas requirements for adaptation and repeat-spacer duplications. (A) Gel images show bands resulting from four rounds of PCR targeting the leader-adjacent end of the CRISPR array; expanded (Ex) and unexpanded (Un) products are indicated. At least three independent experiments were done; images shown here are representative. Gene deletions were designed to disrupt adaptation (Δcas1 and Δcas2 strains), crRNA maturation (Δcas6) or interference (Δcsm1-5 and Δcsm1-6 strains). (B) Transformation efficiencies for a non-target plasmid (pNo-target), a transcribed target plasmid (pTarget), and a non-transcribed target plasmid (pTarget-NT) are shown for four strains (n = 3). (C) Genome-wide view of aligned new type III spacers. The y-axis indicates the cumulative density of aligned spacers counting either all supporting reads (total) or only unique reads, with respect to alignment coordinates (unique). Spacers aligned to the top strand are colored blue; bottom strand are red. The positions of the native type II and type III arrays are indicated in orange; rRNA and tRNA gene positions are show in green. Black bars above the unique spacer track show regions where significant spacer enrichment was detected. (D) Schematic of PCR products corresponding to an unexpanded array, a novel spacer, or duplication of existing spacers in the array. (E and F) Genome browser tracks show the cumulative density of aligned new spacers across the introduced type II-A array (E) and the native type III-A array in WT and Δcas1 strains (F); new spacers that align to existing spacers correspond to spacer duplications. Spacer duplications were observed for all strains examined; representative genome browser tracks for the Δcas1 and WT strains are shown here (F). At least three independent experiments were conducted for each strain and pooled data were used to determine the percent of all expanded reads that corresponded to spacer duplications (“Dup”, rather than novel spacers); that percent is indicated with along with the total number of aligned spacers and replicates.