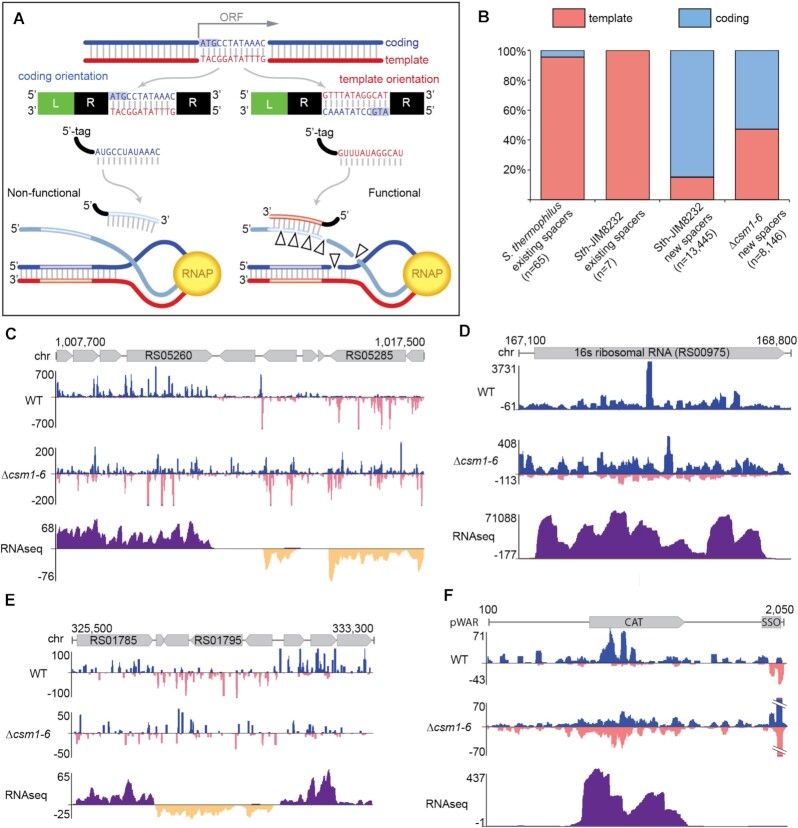
Figure 3.
New type III-A spacers aligned to coding and template strands of expressed genes. (A) Diagram illustrating the outcomes for type III target interference given two possible orientations for spacer integration. When a type III-A spacer corresponds to (aligns to) the coding strand of a gene, the expressed type III-A crRNA will not be able to perform defense (left). In contrast, when a spacer corresponds to the template strand, the expressed type III crRNA can bind nascent RNA and initiate defense (right). (B) Proportions of type III-A spacers derived from the coding strand (blue) and template strand (pink) are shown for existing array spacers and newly acquired, leader-adjacent spacers identified in this work. Existing spacers in type III-A arrays from 63 sequenced S. thermophilus genomes were aligned to a database of phage genomes and for those spacers that aligned, strand was noted (identical spacers were counted once). Sth JIM8232 (WT) existing spacers were aligned similarly. For newly acquired spacers, pooled data of at least three independent experiments are presented. (C–F) Genome browser tracks show cumulative spacer density and RNAseq read density across selected genome and plasmid regions for WT and Δcsm1-6 strains. Spacers aligning to the top strand are colored blue; spacers aligning to the bottom strand are pink. For RNAseq data, RNA reads aligning to the top strand are dark purple and reads aligning to the bottom strand are dark yellow. The y-axis indicates the depth of aligned sequences (spacers or RNAseq reads). For spacers, data from at least three independent experiments was combined on a single track. For RNAseq, aligned data from a single representative replicate is shown.