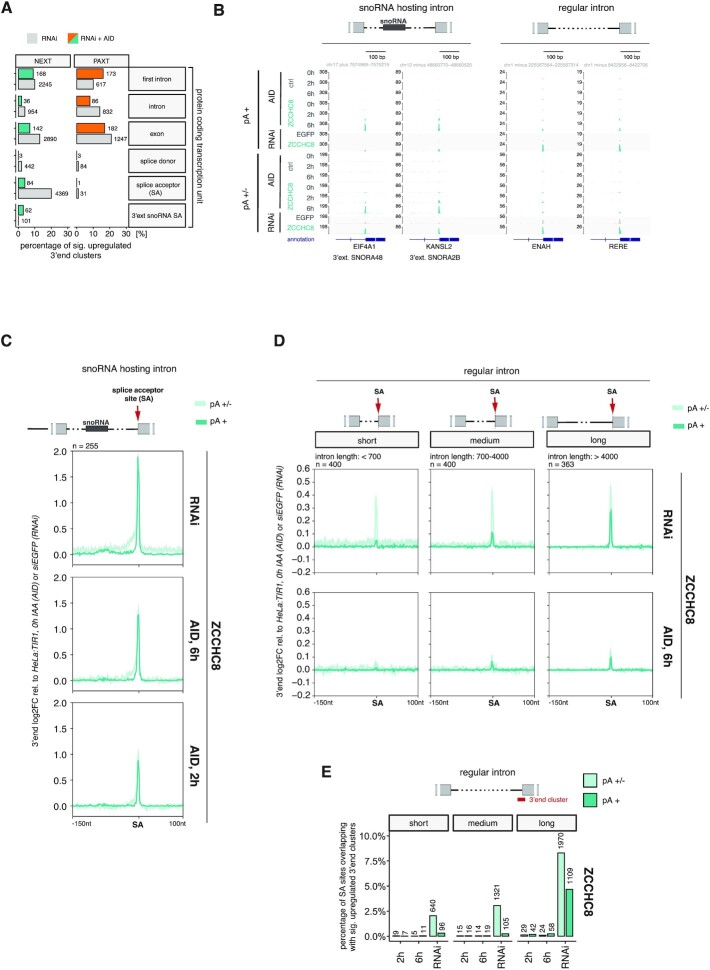
Figure 4.
Rapid ZCCHC8 depletion establishes NEXT targeting of snoRNA-hosting introns. (A) Bar plots showing the percentage of 3′ end clusters within ‘RNAi’- and ‘RNAi + AID’-based NEXT and PAXT targeting classes that overlap with the indicated features within protein coding TUs. Respective absolute numbers of 3′ end clusters are included next to the individual bars. (B) Genome browser views of selected SAs of snoRNA-hosting (left) or regular (right) introns, showing data from AID- (0, 2 and 6 h) and RNAi-mediated ZCCHC8 depletion samples. pA+ and pA+/− libraries are displayed as in Figure 3D. HeLa:TIR1 and siEGFP ctrl samples were included as controls. (C) Metagene profiles of the indicated regions up- and downstream of SA sites of snoRNA-hosting introns (see schematics on top). Values shown are log2FC in single nucleotide resolution between pA+ and pA+/− 3′ end-seq RNAi or 6 h AID libraries following ZCCHC8 depletion relative to their respective controls (RNAi: siEGFP; AID: HeLa:TIR1, 0 h IAA). The displayed region covers 150 nt upstream and 100 nt downstream the SA anchor point. (D) Metagene profiles as in (C), but of regular introns in otherwise snoRNA-hosting transcripts and stratified into three different classes, according to intron length (left: <700 bp; middle: 700–4000 bp; right: >4000 bp). (E) Bar plots showing the percentage of SA sites of short, medium and long introns [see panel (D)], overlapping with significantly upregulated 3′ end clusters in ZCCHC8 depletion conditions (2 h, 6 h, RNAi) relative to the respective controls (AID: 0 h IAA; RNAi: siEGFP). Absolute numbers of SA sites are included on top of the individual bars.