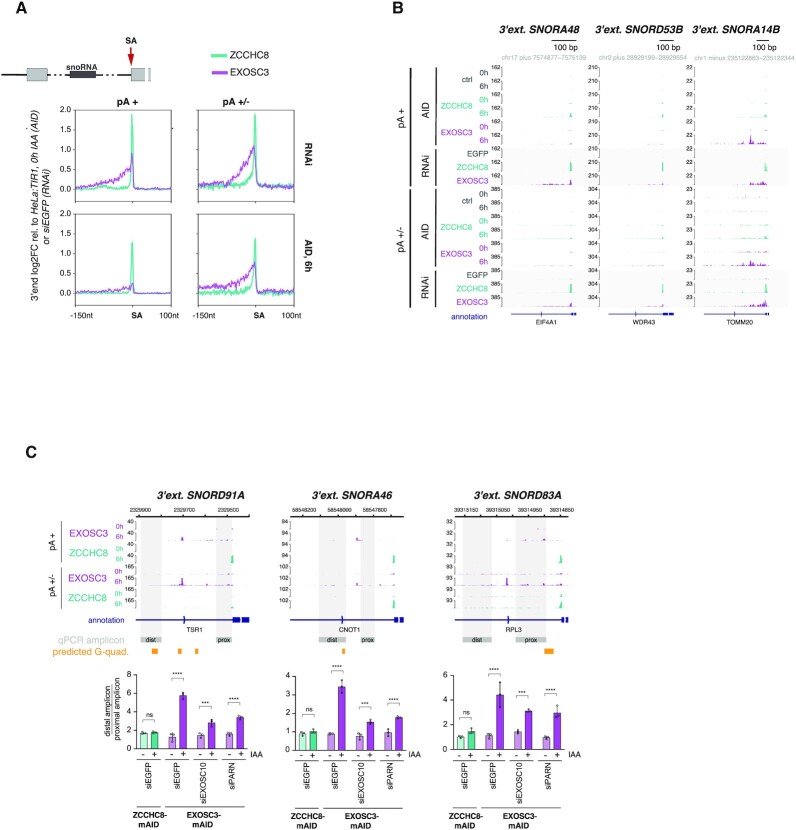
Figure 5.
3′ end trimming in EXOSC3 depletion conditions. (A) Metagene profiles of regions around SA sites of intron-hosted snoRNAs, showing log2FC in single nucleotide resolution between pA+ and pA+/− 3′ end sequencing data of newly synthesized RNA upon RNAi- or 6 h AID-mediated ZCCHC8 and EXOSC3 depletion relative to siEGFP or unedited HeLa:TIR1, 0 h IAA controls. Regions are displayed as in Figure 4C. (B) Genome browser views as in Figure 3D, but showing regions around selected SAs of snoRNA-hosting introns. Shown data are from AID- (0 and 6 h) or RNAi-mediated ZCCHC8 and EXOSC3 depletion conditions. (C) Trimming assay based on RT-qPCR analysis of selected amplicons (gray areas and ‘qPCR amplicon’ annotations in genome browser view) within 3′ extended snoRNAs. RT-qPCR analyses were conducted on total RNA isolated from EXOSC3-mAID (purple) and ZCCHC8-mAID (green) cells treated with siRNAs against EGFP, EXOSC10 or PARN and followed by 0 h (−) or 6 h (+) exposure to IAA. Data were normalized to Rplp0 mRNA (RPO) and displayed as the trimming factor, obtained from the ratio of distal/proximal amplicon values. Differences within RNAi treatment groups, comparing +/− IAA samples, were determined by a two-way ANOVA test [*Padj ≤ 0.05, **Padj ≤ 0.01, ***Padj ≤ 0.001, ****Padj ≤ 0.0001, nonsignificant (ns) = Padj > 0.05].