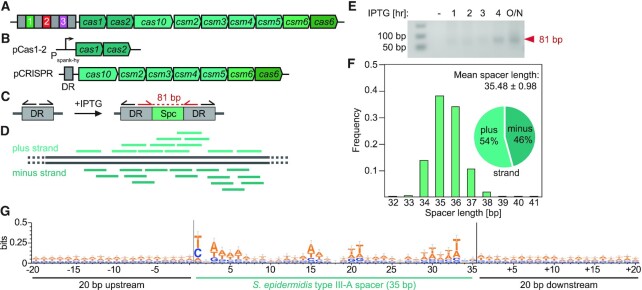
Figure 1.
An inducible system for studying spacer acquisition by the S. epidermidis type III-A CRISPR-Cas system. (A) Schematic representation of the S. epidermidis type III-A CRISPR-Cas system. (B) Schematic representation of the two-plasmid system used in the inducible adaptation assay for S. epidermidis type III-A CRISPR-Cas. (C) Sensitive DR-PCR design with repeat-annealing primers (arrows) that allows for detection of rare acquisition evens. (D) After next-generation sequencing of DR-PCR products and extraction of spacer sequences, spacers are mapped to the bacterial genome. (E) Agarose gel electrophoresis of DR-PCR products sampled at different time points. (F) Spacers acquired 3 h post induction of the S. epidermidis type III-A acquisition machinery are 35.48 ± 0.98 (mean ± SD) bp long. 54% and 46% match the plus and minus strand of the bacterial genome, respectively. (G) Weblogo analysis of genome-aligned spacers acquired by the S. epidermidis type III-A CRISPR-Cas system, along with 20 bp upstream and downstream. The y-axis was modified (from the default of 2 bits to 0.5 bit) to allow for better resolution, hence the height of the letters does not represent a significantly enriched motif. Only 35 bp long spacers were included in this alignment, n = 1460.