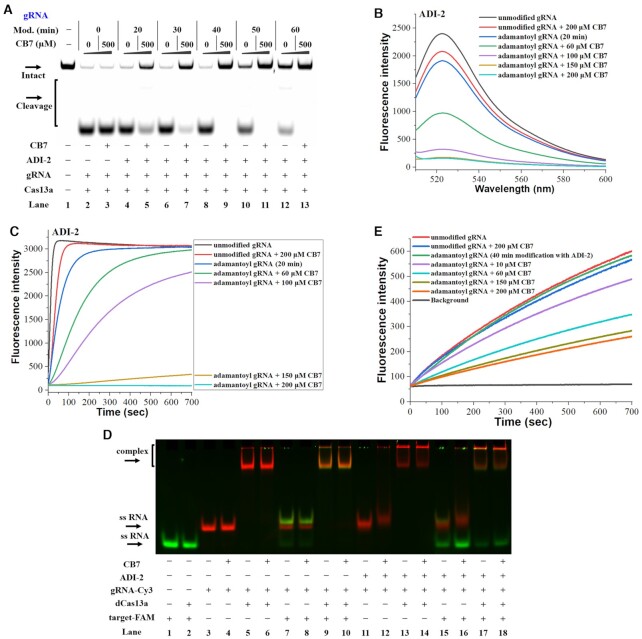
Figure 4.
Supramolecular OFF-switches for CRISPR/Cas13a. Reactions were performed as described in the Materials and Methods section. All samples were tested in three biological replicates. Image of representative data was shown here. (A) Modification-dependent inhibition of CRISPR/Cas13a upon supramolecular complexation. In this study, adamantoyl gRNAs and Cas13a were employed to cleave target1 in the absence or presence of 500 μM CB7. Lane 1: no enzyme control; lanes 2, 4, 6, 8, 10, 12: no CB7 control; lanes 3, 5, 7, 9, 11, 13: the 500 μM CB7 treatments. Lanes 2–3 contain unmodified gRNA; lanes 4–5, 6–7, 8–9, 10–11, 12–13 contain adamantoyl gRNAs with increasing modification levels. (B) The influence of supramolecular complexation on the Cas13a collateral cleavage. The fluorescence spectra were measured at room temperature after 100 s incubation at 37°C. (C) Dose-dependent effect of supramolecular complexation on inhibiting CRISPR/Cas13a. (D) Effect of supramolecular complexation on the formation of ternary dCas13a-gRNA-target complexes. Lanes 1–2: target-FAM control; lanes 3–10 contain unmodified gRNA-Cy3; lanes 11–18 contain adamantoyl gRNA-Cy3 (a 60 min modification with 100 mM ADI-2). (E) Effect of supramolecular complexation on RNA hybridization. This study uses adamantoyl gRNAs (a 40 min modification with 100 mM ADI-2).