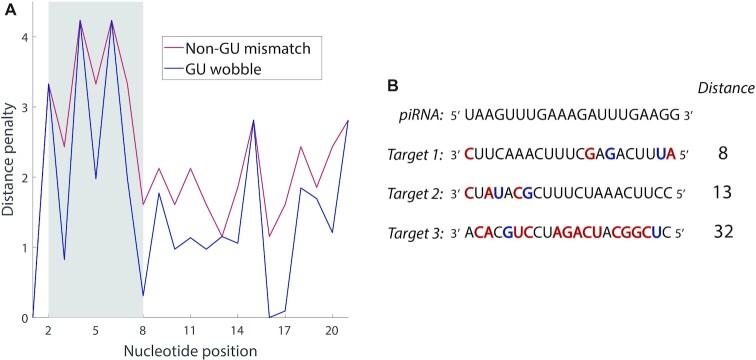
Figure 2.
Overview of the functional piRNA distance metric. (A) The penalty associated with a non-GU mismatch or GU wobble on each nucleotide of potential piRNA-target pairs. The gray shaded region shows the seed region of the piRNA, which has higher penalties than elsewhere. (B) Example piRNA-target distances for a piRNA and three potential targets, with mismatches shown in red and GU wobbles shown in blue. Target 1 is a ‘typical’ actual target for the piRNA. Target 2 possesses the same number of mismatches as Target 1, but they have been relocated to the seed region. Target 3 has roughly the number of mismatches and wobbles that a random 21-nt sequence would possess.