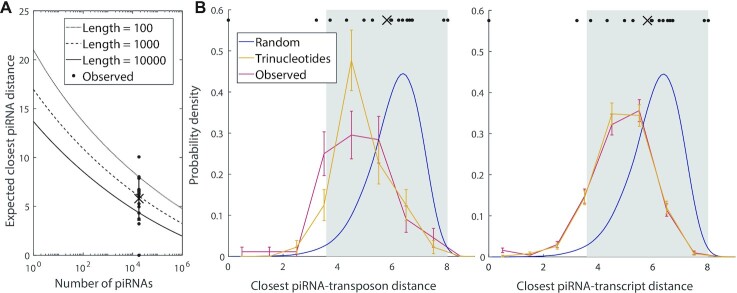
Figure 5.
Comparison of actual global targeting by all piRNAs to the random global closest-match distribution of piRNA-target distances (see Materials and Methods). (A) Predicted mean distance for the closest match between a ‘target’ (random sequence of length L = 100, 1000 or 10 000), and any of the ‘piRNAs’ (random 21 nt sequences) as a function of the number of distinct ‘piRNAs’. The data points plotted above the true number of C. elegans piRNAs (17 849) show the distances for the 17 actual piRNA-target pairs studied by Zhang et al. (5), with the cross and error bar indicating the mean and standard deviation. (B) Probability distribution of global closest-match distribution for actual transposons (left) and self-transcripts (right), of similar lengths, with real and random piRNAs. Red curves represent data for real piRNAs (n = 17 849) targeting 800–1200 nt C. elegans genes, either transposons (n = 90, left) or a random sample of self-transcripts (n = 500, right). Yellow curves represent targeting of the same transposons and transcripts by randomly generated piRNAs with the same position-specific trinucleotide probabilities as real piRNAs. Blue curves show the smoothed probability density of closest distances for 17 849 fully random ‘piRNAs’ with a fully random ‘gene’ of 1000 nt. Error bars indicate counting error. The data points plotted above the probability distributions show the distances for the 17 actual piRNA-target pairs presented in (A), while the cross and gray shaded region show the mean and standard deviation, respectively.